Fig. 3. htFuncLib exposes a large space of functional multipoint active-site GFP variants.
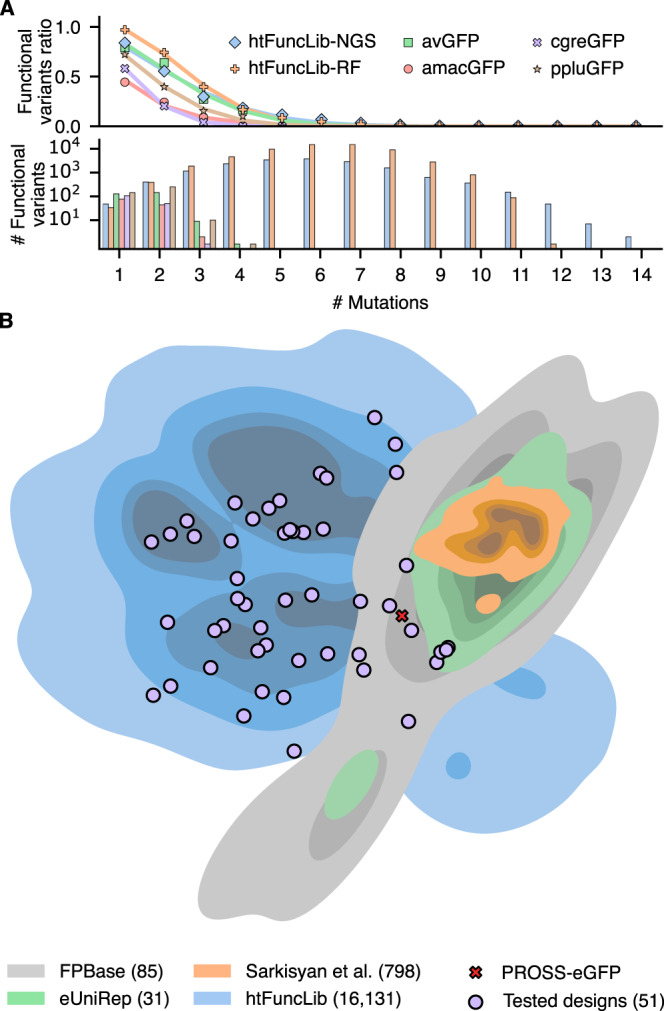
Deep sequencing of htFuncLib libraries sorted by fluorescence revealed over 16,000 potentially active designs. A Frequency and number of functional variants with a given number of mutations (top and bottom, respectively). htFuncLib-NGS - all sequences obtained from deep sequencing of the sorted designs; htFuncLib-RF - the entire sequence space labeled by the random forest. The avGFP dataset was derived from Sarkisyan et al.4. The amacGFP, cgreGFP, and ppluGFP datasets were derived from Somermeyer et al.3. Lines represent fits to the data (points) according to Eq. 2 (see “Methods” and Supplementary Table 2). Data excluded sequences with mutations outside of the chromophore pocket. B Distance-preserving dimensionality reduction analysis shows the relationships between GFP variants in FPBase35, Sarkisyan et al.4, eUniRep41, and htFuncLib. The plot approximates the number of mutations between any pair of mutants41,75. PROSS-eGFP (and eGFP, which are nearly identical in the designed positions, Supplementary Dataset 7) are marked by a cross for reference. Individually characterized htFuncLib designs are marked by purple circles. The number of sequences represented for each category is marked in parentheses. Variants with mutations outside the chromophore pocket were included, but these mutations were ignored when calculating distances. Data are provided as a Source Data file.
