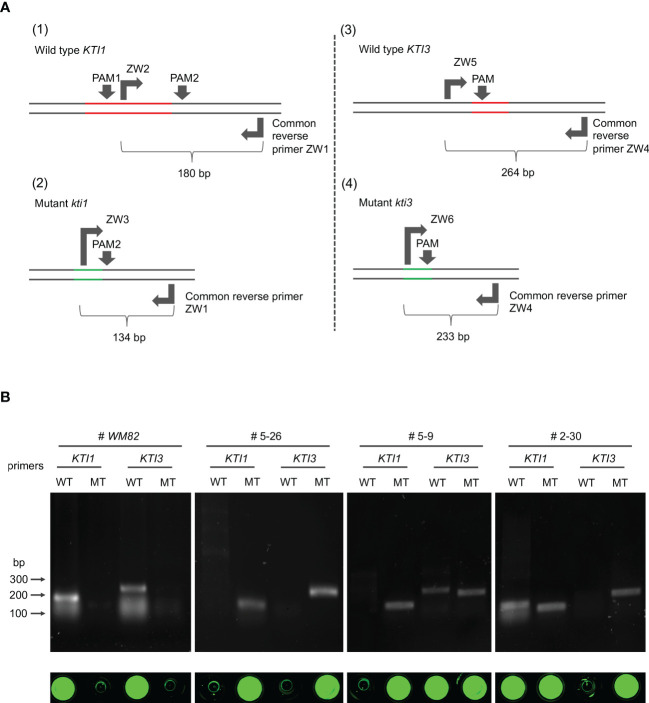
Figure 6.
The development of selection markers for breeding low KTI soybean varieties based on the kti1 and kti3 mutants generated by CRISPR/Cas9-mediated gene editing. (A) Schematic development of primers for amplification of wild type KTI1 (1), mutant KTI1 (2), wild type KTI3 (3), and mutant KTI3 (4). The red lines indicate the lost fragment in KTI1 or KTI3 during gene editing. The green lines indicate new DNA regions in kti1 or kti3 generated by splicing two fragments. (B) The 4 pairs of primers in (A) were utilized to amplify the alleles of KTI1, kti1, KTI3, and kti3 with gDNA of four different soybean genotypes, including WM82, three transgenic lines #5-26, #5-9, and #2-30. Based on our genotyping data, #5-26 has homozygous mutations of kti1 and kti3; #5-9 only has a homozygous mutation of kti1 but carries the heterozygous mutation of kti3; #2-30 only has a homozygous mutation of kti3 but carries the heterozygous mutation of kti1. Thus, it was clear that the pair of ZW1/ZW2 can amplify wild type KTI1 from WM82 and #2-30 gDNA in PCR tests, while the pair of ZW1/ZW3 can amplify mutant kti1 from #5-9 and #5-26 gDNA. Also, the pair of ZW4/ZW5 can amplify wild type KTI3 from WM82 and #5-9 gDNA, while ZW4/ZW6 can amplify mutant kti3 from #2-30 and #5-26 gDNA. As shown in the bottom panel, only the positive PCR products incubated with the dye of sybrgreen at 75°C can display the fluorescent signals, suggesting the reliability of the developed gel-electrophoresis-free method for screening mutant alleles of kti1 and kti3.