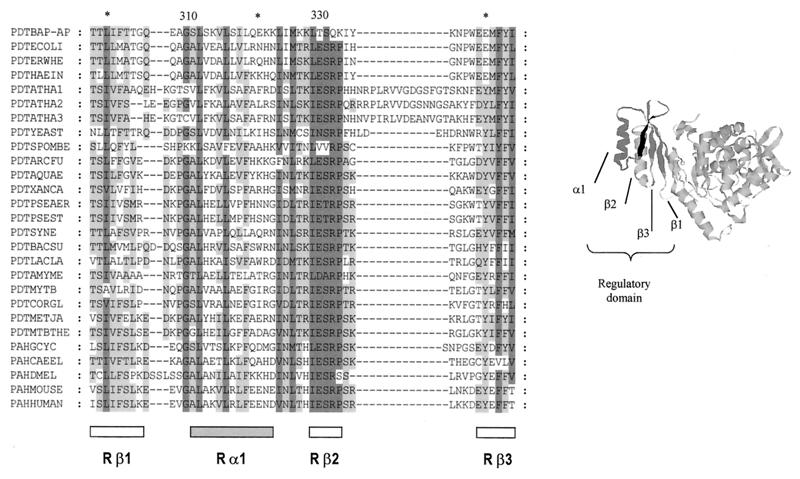
FIG. 2.
Alignment of the homologous region of the regulatory domains from PDT and PAH proteins. α-Helix and β-strand positions correspond to the information obtained from the crystal structure of rat PAH (5) (shown to the right). The sequences used in this alignment were as follows (sources and accession numbers are given in parentheses): PDTBAP-AP (B. aphidicola [A. pisum], AJ239043), PDTECOLI (E. coli, P07022), PDTERWHE (E. herbicola, Q02286), PDTHAEIN (Haemophilus influenzae, P43900), PDTATHA1 (Arabidopsis thaliana, O22241), PDTATHA2 (A. thaliana, AAD30242), PDTATHA3 (A. thaliana, AAC73018), PDTYEAST (Saccharomyces cerevisiae, P32452), PDTSPOMBE (Schizosaccharomyces pombe, O14361), PDTARCFU (Archaeoglobus fulgidus, O30012), PDTAQUAE (Aquifex aeolicus, O67085), PDTXANCA (Xanthomonas campestris, O87954), PDTPSEAER (P. aeruginosa, Pseudomonas Genome Project), PDTPSEST (P. stutzeri, P27603), PDTSYNE (Synechocystis sp., P72808), PDTBACSU (Bacillus subtilis, P21203), PDTLACLA (Lactococcus lactis, P43909), PDTAMYME (Amycolatopsis methanolica, Q44104), PDTMYTB (Mycobacterium tuberculosis, P96240), PDTCORGL (Corynebacterium glutamicum, P10341), PDTMETJA (Methanococcus jannaschii, Q58054), PDTMTBTHE (Methanobacterium thermoautotrophicum, 027288), PAHGCYC (Geodia cydonium, Y16353), PAHCAEEL (Caenorhabditis elegans, CAA91286), PAHDMEL (Drosophila melanogaster, Q27599), PAHMOUSE (Mus musculus, P16331), PAHHUMAN (Homo sapiens, P00439). The numbers at the top indicate the residue position in Buchnera.