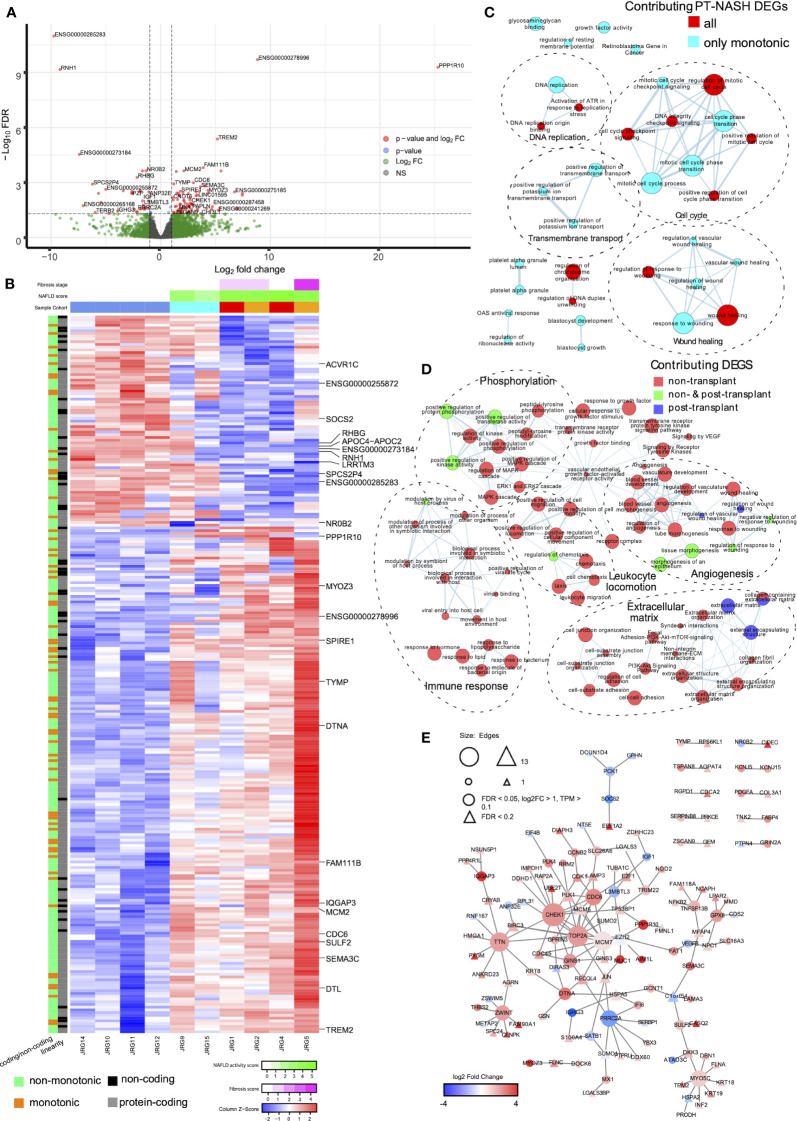
Figure 1.
Gene expression patterns, enriched pathways, and protein-protein interaction (PPI) networks in livers with post-transplant and non-transplant NASH. (A) Volcano plot of differential gene expression in PT-NASH compared to control. Green: log2 FC ≥ 1; blue: FDR ≤ 0.05; red: log2 FC ≥ 1 and FDR ≤ 0.05. (B) Heatmap of TPM values of all PT-NASH DEGs. Sample annotations are shown at the top. The first row represents fibrosis score (purple), the second row represents NAFLD activity score (NAS, green); and the third row represents sample group (blue: control; cyan: steatosis; orange: de novo NASH; red: recurrent NASH). Gene annotations are shown on the left (orange: monotonic; green: non-monotonic; grey: protein-coding genes; black: lncRNAs). The 25 top DEGs are labelled on the right. (C) Enrichment map of pathways found in monotonic and non-monotonic PT-NASH expression patterns (ActivePathways, FDR < 0.2). Each pathway is a node, edges connect pathways sharing many genes, and colors indicate DEGs contribution to pathway enrichments (cyan: monotonic; red: combined monotonic and non-monotonic). Subnetworks were annotated manually to combine similar pathways (dashed circles). (D) Map of enriched pathways in PT-NASH and NT-NASH (ActivePathways, FDR < 0.2). Colors indicate disease type contributing to pathway enrichment (red: NT-NASH; blue: PT-NASH; green: combined). (E) PPI network interactions with DEGs in PT-NASH. Color indicates expression fold-change (PT-NASH vs. control livers), and circles show the most significant DEGs. Triangles indicate additional less-significant genes (FDR < 0.2).