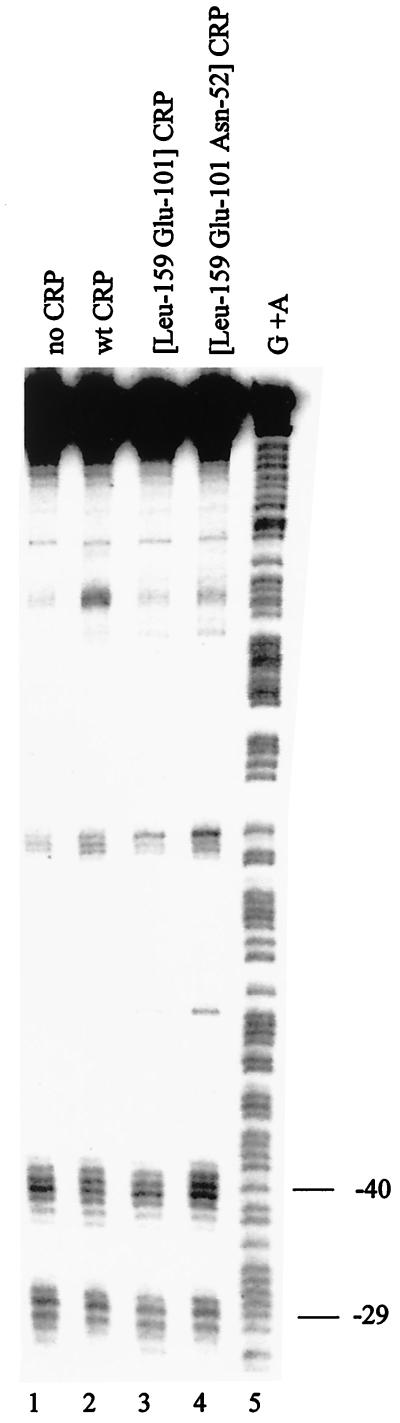
FIG. 2.
DNA cleavage of galp19T by RNAP containing FeBABE-tagged [Cys581] ς70. Purified wild-type (wt) CRP (150 nM) or mutant derivatives were mixed, as indicated, with 32P-end-labeled AatII-HindIII DNA fragments (0.4 nM, labeled on the template strand) carrying the gal promoter region with the p19T mutation in a reaction volume of 35 μl [20 mM HEPES (pH 8.0), 5 mM MgCl2, 200 μM cyclic AMP, 50 mM potassium glutamate, 50 μg of bovine serum albumin per ml, 5 μg of poly(dI-dC)]. RNAP holoenzyme (30 nM) reconstituted with FeBABE-tagged [Cys-581] ς70 was then added and incubated at 37°C for 20 min. Complexes were challenged with heparin (200 μM for 5 min), and DNA cleavage was initiated by the addition of sodium ascorbate (2 mM) followed by incubation at 37°C for 20 min. Modified DNA was extracted with phenol-chloroform and analyzed on a 6% polyacrylamide sequencing gel, which was calibrated with Maxam-Gilbert G+A sequence ladders and processed and scanned using a Molecular Dynamics PhosphorImager (full protocols are given in reference 3). Bands due to DNA cleavage near positions −40 and −29 upstream of the galp1 transcription start point are indicated. Lane 1, no CRP; lane 2, wild-type CRP; lane 3, [Leu159 Glu101] CRP; lane 4, [Leu159 Glu101 Asn52] CRP; lane 5, G+A sequencing ladder.