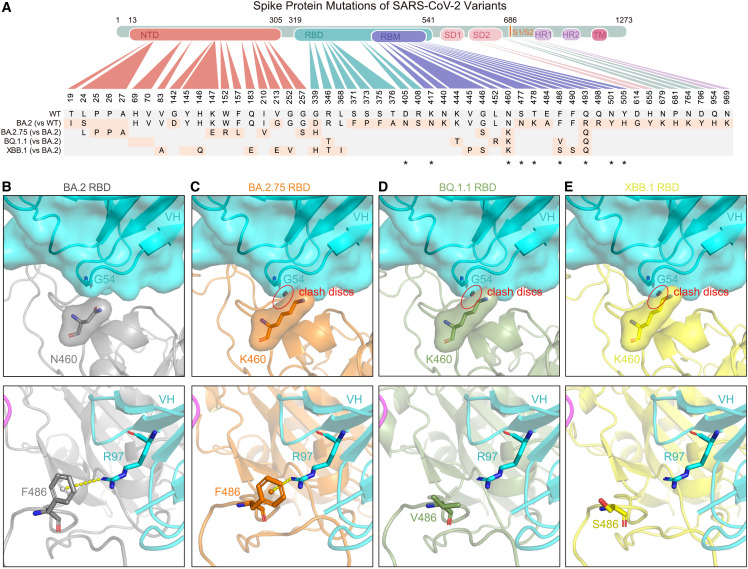
Figure 3.
Sequence alignment and structural analysis of Omicron BA.2.75, BQ.1.1, and XBB.1 subvariants
(A) Key mutations in the spike protein of Omicron BA.2, BA.2.75, BQ.1.1, and XBB.1. “∗” represents mutations appeared in the binding epitopes of VacBB-551.
(B–E) Local region of N460 and F486 of BA.2 RBD (B), K460 and F486 of BA.2.75 RBD (C), K460 and V486 of BQ.1.1 RBD (D), and K460 and S486 of XBB.1 RBD (E) with G54 and R97 of VacBB-551 heavy chain, respectively. The structures of BA.2.75, BQ.1.1, and XBB.1 RBDs were predicted by AlphaFold2 (https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb). The clash discs were displayed using the “Show bumps” plugin (https://pymolwiki.org/index.php/Show_bumps) in Pymol (C–E). The pseudoatom of F486 is shown as nb_sphere and the cation-π interaction is shown in a yellow dashed line (B and C). N460, K460, F486, V486, S486, G54, and R97 residues were shown as sticks. The VacBB-551 heavy chain was shown as cartoon/surface colored by cyan. RBDs of BA.2, BA.2.75, BQ.1.1, and XBB.1 are shown in gray, orange, smudge, and yellow, respectively.
See also Figure S5.