Figure 2. HIV-1 in CSF is replicating and adapted to growth in T cells.
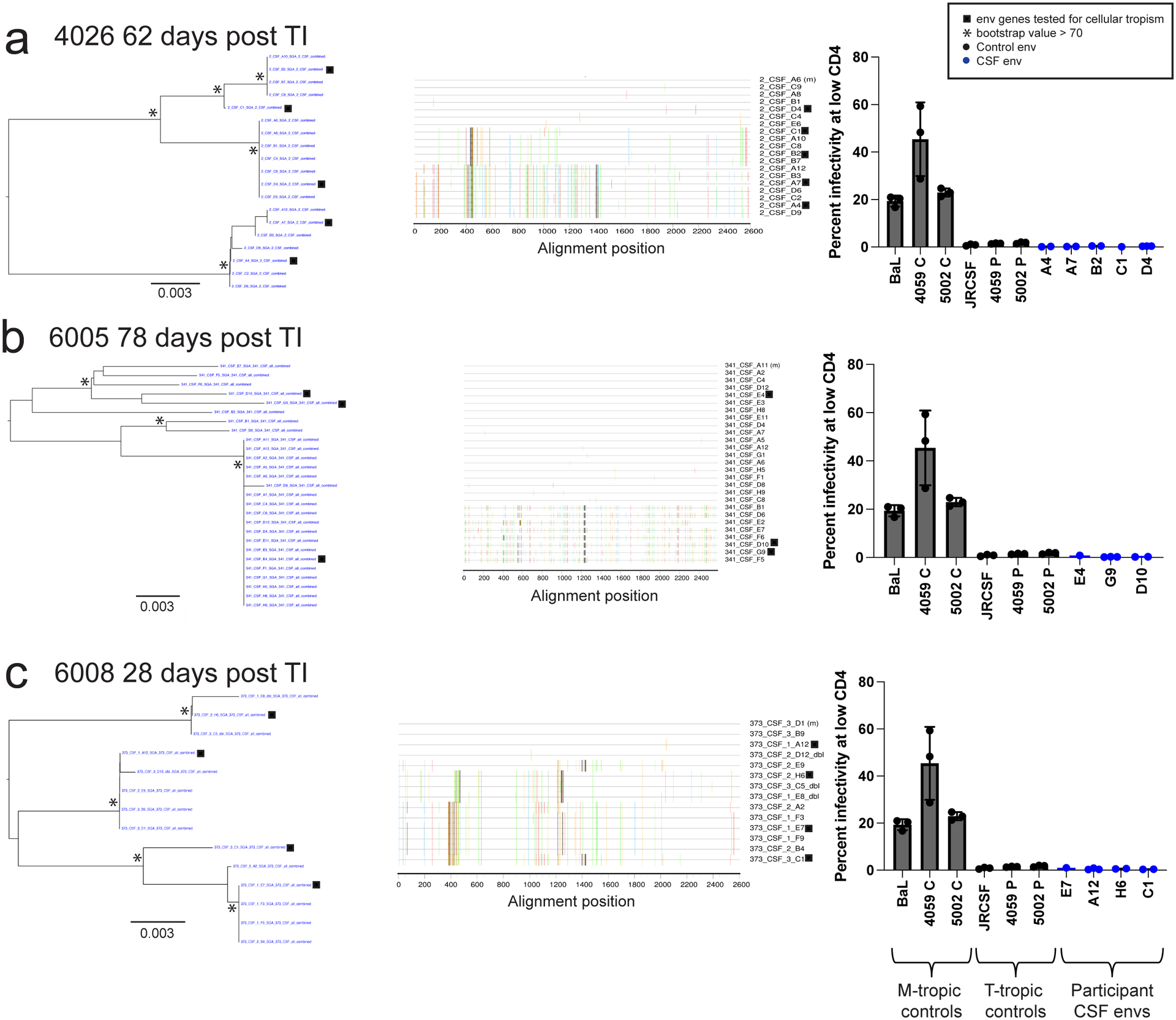
Full length env gene amplicons were generated by end-point dilution PCR of cDNA synthesized using viral RNA in the CSF as template. The entry phenotype was assessed for three representative participants with clonal CSF-specific lineages in the viral rebound populations following TI at the indicated time: 4026 (a), 6005 (b), and 6008 (c). Panel 1 shows neighbor-joining phylogenetic trees of full-length env gene sequences. Panel 2 to the right is a highlighter plot of the full-length env gene sequences depicting the low but detectable sequence diversity within these lineages in each participant. The third panel (far right) graphs the relative infectivity at low levels of CD4 for macrophage-tropic env gene controls used to pseudotype a reporter virus, T cell-tropic env gene controls, as well as the participant-derived env genes shown in the first panel. 3 technical replicates were done for each clone and 1–3 replicate clones were tested for each env. Specific numbers of clones tested for each env are as follows; A4: 2 clones, A7: 2 clones, B2: 2 clones, C1: 1 clones, D4: 3 clones, E4: 1 clones, G9: 3 clones, D10: 2 clones, E7: 1 clone, A12: 3 clones, H6: 2 clones, C1: 2 clones. The infectivity at low CD4 values for each replicate clone are shown with the dots on the bar graph and the mean and standard deviation values for the replicates are shown with the bar and error bars, respectively. All env genes cloned from the CSF virus of the three participants were T cell-tropic when used to pseudotype the reporter virus and tested in the CD4lowCCR5high/CD4highCCR5high Affinofile cell entry assay.
