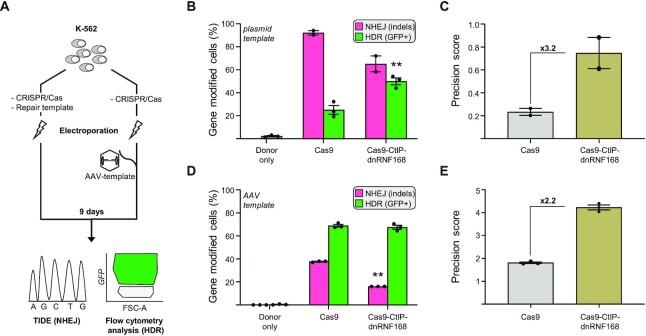
Figure 3.
CRISPR/Cas9-CtIP-dnRNF168 promotes the targeted integration of large expression cassettes. (A) Experimental design. K562 cells were electroporated with the corresponding CRISPR/Cas9 expression plasmids together with a plasmid as repair template (left) or transduced immediately upon electroporation with an adeno associated virus (AAV)-based vector containing the HDR template (right). Nine days later, NHEJ and HDR frequencies were measured via TIDE or flow cytometry, respectively. (B, D) The graphs show the frequency of gene edited K562 cells that underwent either NHEJ- or HDR-mediated DSB resolution, measured by TIDE (NHEJ) or flow cytometry (HDR), respectively. (C, E) Graphs show the precision score, calculated as the ratio of HDR to NHEJ events of the results shown in panels B and D, respectively. Fold change, as compared to the unmodified CRISPR/Cas9 nuclease, is indicated within the graph. Each dot represents biological replicates. Statistically significant differences, as compared to the unmodified CRISPR/Cas9 nuclease, are indicated with asterisks within the graphs and correspond to P-values calculated with a two-tailed, paired Student's t-test (**P< 0.01). Error bars indicate SEM.