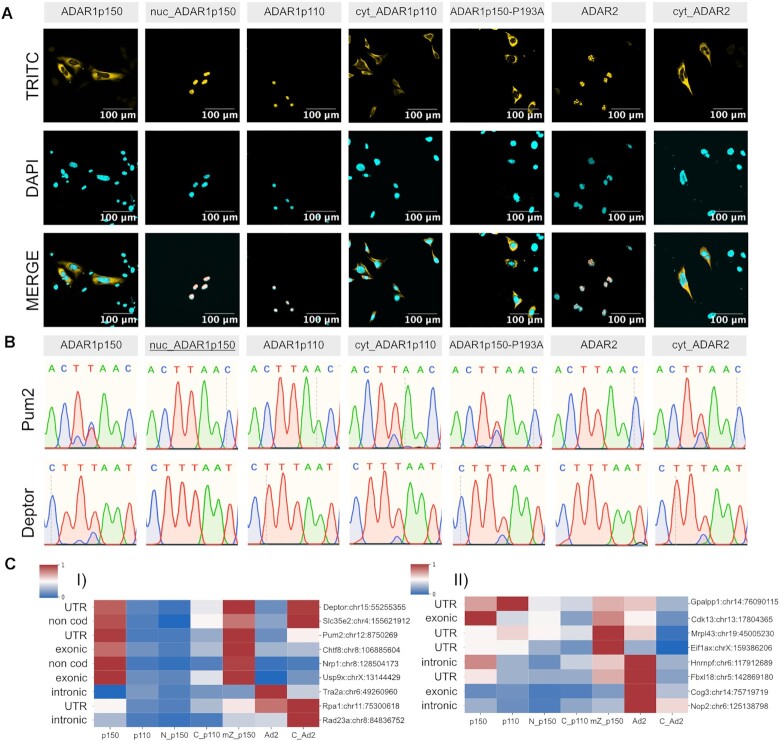
Figure 2.
ADAR1-isoform specificity is partially driven by its cellular localization. (A) Confocal microscopy images confirm the cellular localization of mislocalized ADAR mutants. A P193A mutation in ZBDα remains cytoplasmic. TRITC channel shows transfected constructs in confocal sections. DNA is stained with DAPI (scale bar: 100 μm). (B) Sanger sequencing traces showing the impact of mislocalization and ZBDα mutation on editing-specificity of ADAR1. Pum2 (chr12:8750269) and Deptor (chr15:55255355) are efficiently edited by wild-type ADAR1p150 but not by wild-type ADAR1p110. Cytoplasmic ADAR1p110 and cytoplasmic ADAR2 also edit both selected targets, however, only at one of the two adjacent sites. In contrast, nuclear ADAR1p150 does not show any editing of selected targets. The P193A mutation does not affect Deptor editing but leads to reduced editing at one of the two sites in Pum2. The reverse strand is sequenced showing T to C conversion in the chromatogram. (C) Heat maps of editing by wild-type and mutated ADAR versions at selected editing sites detected by amplicon-seq. I) Impact of mislocalization on editing of ADAR1p150 targets. II) Impact of mislocalization on editing of ADAR1p110 targets. At ADAR1p150 sites, the P193A mutation does not affect editing but nuclear localization of ADAR1p150 reduces editing. At the same time, cytoplasmic localization of ADAR1p110 or ADAR2 allows editing of ADAR1p150 sites (p150 = ADAR1p150, p110 = ADAR1p110, N_p150 = nuclear ADAR1p150, C_p110 = cytoplasmic ADAR1_p110, mZ_p150 = mutant ZBD in ADAR1p150, Ad2 = ADAR2, C_Ad2 = cytoplasmic ADAR2). Names and chromosomal positions of editing sites are indicated at the right of the heat map.