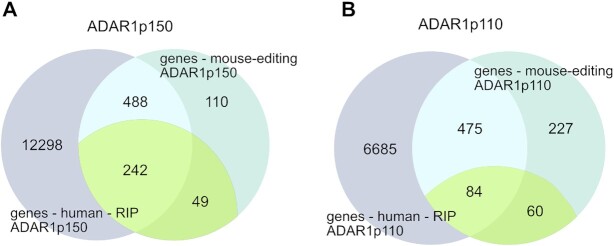
Figure 4.
Intersection of regions bound by ADAR1 isoforms identified by ADAR1 RIP-seq in human cells with editing regions identified in mouse genes. (A) More than 80% (242) of the genes containing non-repetitive editing sites were detected in both experiments performed with ADAR1p150. (B) Only ∼59% (84) was identified between two experimental approaches for ADAR1p110. In purple: genes that were identified by RIP-seq in human cells; turquoise: genes that were identified as editing targets in MEFs; yellow-green: genes containing editing sites in non-repetitive regions and those that were edited in MEFs.