Figure 1.
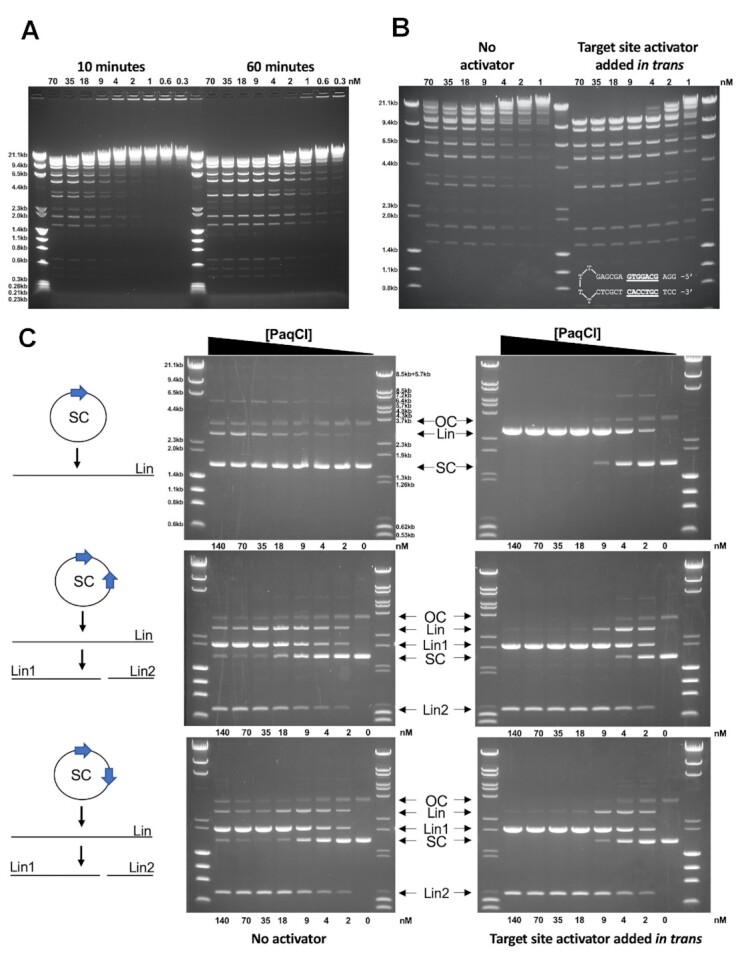
Cleavage activity of PaqCI towards lambda phage DNA and plasmid substrates. (A) PaqCI demonstrates optimal activity at an equal to modest excess of enzyme binding sites to substrate target motifs, with an excess of enzyme to substrate leading to faster, but not more complete, cutting. Lambda phage DNA, with 12 PaqCI sites, are digested and quenched at various time points. Time points correspond to 10- and 60-min digests, with the concentration of enzyme monomers varied from 0.3 to 70 nM. The DNA recognition site concentration in all digests was 7.2 nM. At 60 min, enzyme concentrations greater than 1:1 enzyme-to-sites exhibit near-complete cleavage, while 0.5:1 enzyme-to-sites generates only slightly less cutting, and lesser amounts of enzyme result in more partial digestion. The molecular weight ladders Lambda-HindIII/PhiX174_HaeIII were used in panel a and b which spans 0.23 to 23.2 kb. This experiment was performed once. Concentrations are rounded to the nearest decimal place. (B) PaqCI cleaves DNA more efficiently when a short hairpin DNA duplex (inset in right gel panel of B) containing the enzyme's target site (underlined bases), is added to the reaction as a trans-activating factor. Lambda phage DNA was digested with variable concentrations of PaqCI from 1 to 70 nM (monomeric concentration) in the absence or presence of the trans activator (which was added at a 5:1 ratio to the PaqCI enzyme, i.e. 350 to 5 nM activator). All lanes correspond to 60-min digests. This experiment was performed once. Units were rounded to the nearest decimal place. (C) Further experiments examine the cleavage of plasmid substrates and demonstrate that PaqCI requires the interaction between multiple bound sites for cleavage. All lanes correspond to 60-min digests at fixed DNA concentrations, with variable enzyme (monomeric concentrations). The single-site substrate (11.4 nM sites) is cleaved far less efficiently than the 2-site substrates (22.8 nM sites). The difference in cleavage efficiency between single- or multiple-target plasmid substrates is eliminated by the presence of the activator DNA (5:1 ratio to enzyme) containing a target site added in trans. The key shows open circle (OC), single-cut linear (Lin), double-cut products (Lin1, Lin2), and super-coiled (SC) species. Two molecular weight ladders were used in these experiments: (i) Lambda-HindIII/PhiX174_HaeIII which spans 0.6 kb to 23.1 kb, and (ii) Lambda-BstEII/pBR322-MspII which spans 0.5–8.5 kb. This experiment was performed once.
