Figure 1.
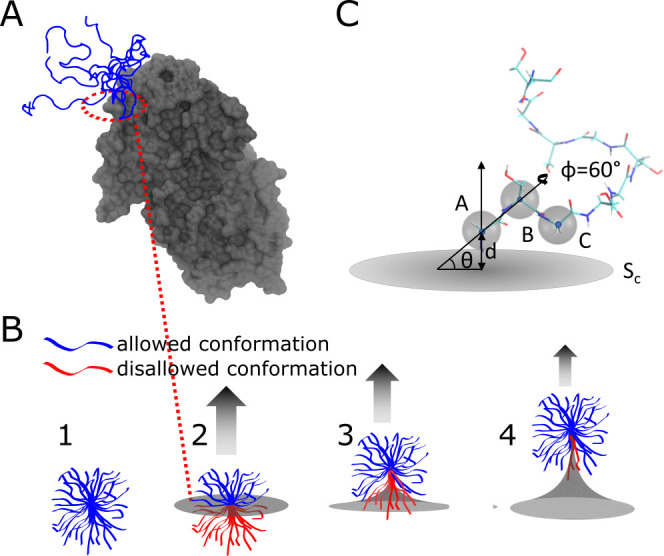
Dynamic IDR conformational ensemble generates an entropic force. (A) IDR tethered to a well-folded domain. Here, the C-terminal IDR tail of the UDP-glucose 6-dehydrogenase (UGDH) protein is shown in blue (with five overlapping conformations to illustrate the variability in the ensemble) tethered to the main folded domain of the enzyme (in gray). PDB obtained from Alphafold232 (https://alphafold.ebi.ac.uk/entry/O60701, accessed Feb. 2023). (B) Schematic showing how a constraining surface alters the conformational entropy of an IDR ensemble. (1) A few representative conformations from an IDR ensemble (blue) occupy an extended volume. (2) As the ensemble is tethered at the terminal to a surface (gray), some conformations clash with the surface (colored in red), causing them to be disallowed and lowering the conformational entropy. (3 and 4) The number of accessible tethered states (ΩT) can be regained by “pulling up” against and pinching the surface (arrow). The ratio between the allowed and total number of conformations for a given ensemble is proportional to the entropic force strength (see eq 3). (C) Enhanced conformational sampling. All conformations of an IDR are aligned along the vector AB connecting the first two Cα atoms. The distance d between the constraining surface Sc and point A is varied to represent tether flexibility. The angle between vector AB and the constraining surface, θ, is varied to represent one degree of rotation for the ensemble, and a second angle, ϕ, represents the rotation angle along the AB vector.
