FIGURE 1. Viral RNA 5′-cap.
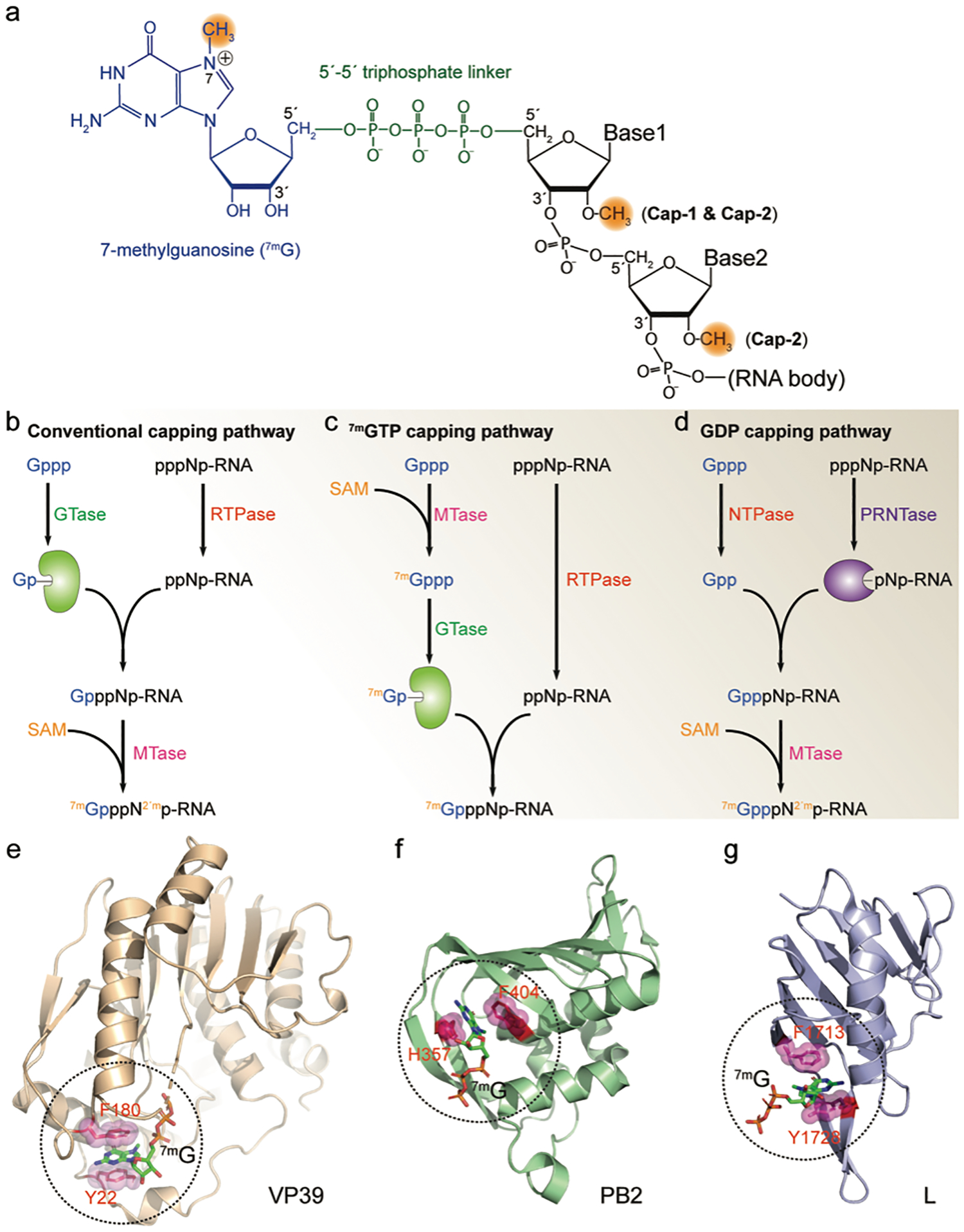
(a) Structure of the 5′-cap of viral mRNA. A 7-methylguanosine (blue) is linked to the RNA 5′-end through a 5′−5′ triphosphate linker (green). The methylations on the N7 and 2′-O are highlighted in orange. (b-d) Viral RNA capping mechanisms. All capping pathways start with GTP (Gppp) and the nascent RNA (pppNp-RNA). GTase, guanylyltransferase; RTPase, RNA triphosphatase; MTase, methyltransferase; SAM, S-Adenosyl methionine; NTPase, nucleoside triphosphatase; PRNTase, polyribonucleotidyl transferase; (e-g) Viral cap binding proteins and their cap-recognition mechanisms. In each protein, two residues with aromatic ring sandwich the 7-methylguanosine cap (7mG) through cation-π interactions. Additional hydrogen bonds further stabilize the protein/cap complex (not shown in the figure). VP39, vaccinia virus MTase (PDB ID 1AV6); PB2, influenza virus polymerase subunit (PDB ID 4C4B); L, Bunyavirus multifunctional polymerase (PDB ID 6QHG).
