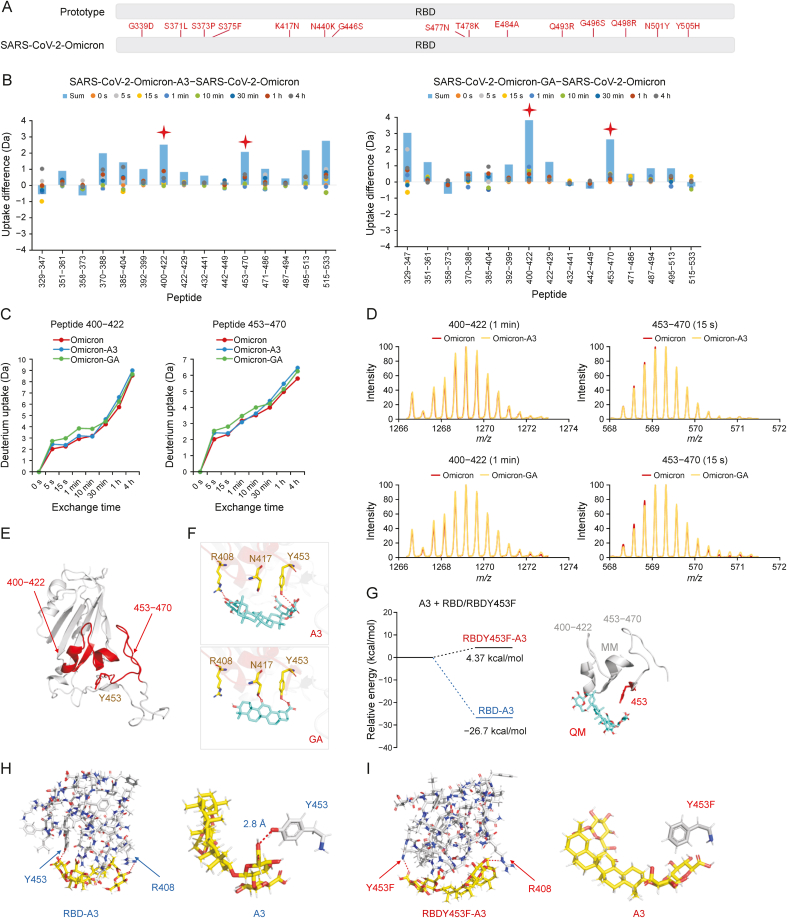
Fig. 3.
Binding mechanisms of licorice-saponin A3 (A3) and glycyrrhetinic acid (GA) targeting the S receptor-binding domain (RBD) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron BA.1. (A) 15 mutations of Omicron S RBD compared to wild type SARS-CoV-2. (B) Deuterium uptake differences between Omicron RBD, and Omicron RBD with A3 or GA. Sum is total uptakes from 0 s to 4 h. (C) Deuterium uptake plots of peptides 400–422 and 453–470 of Omicron RBD, Omicron RBD with A3, and Omicron RBD with GA. (D) Mass spectra of peptides 400–422 and 453–470 of Omicron RBD and Omicron RBD with A3/GA. (E) Location of peptides 400–422 and 453–470 in Omicron RBD. (F) Molecular docking of A3 and GA with Omicron RBD, respectively. Hydrogen bonds (red dashes) are shown. (G) Relative energies of A3 with RBD (peptides 400–422 and 453–470), and A3 with RBDY453F (peptides 400–422 and 453–470) computed by quantum mechanics/molecular mechanics (QM/MM). (H,I) Binding modes of A3 with active residues of RBD (H) and RBDY453F (I), respectively. Hydrogen bonds (red dashes) are shown.