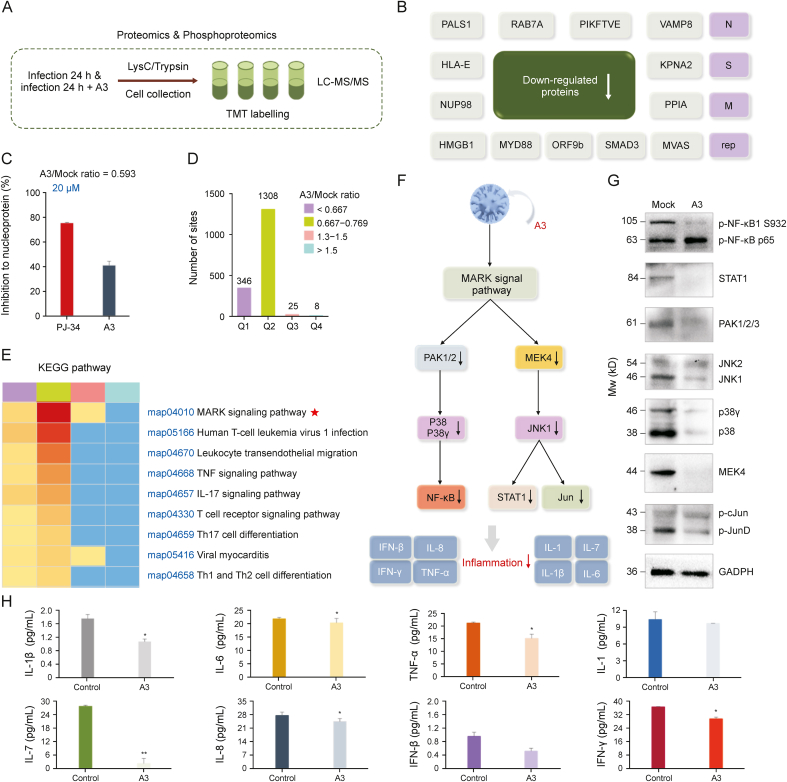
Fig. 4.
Phosphoproteomics profiling of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-infected cells treated with licorice-saponin A3 (A3). (A) Experimental scheme. Vero E6 cells were infected with SARS-CoV-2 (MOI = 1) in the presence or absence of 10 μM A3 for 24 h. Proteins were prepared using lysis buffer (8 M urea, 1% protease inhibitor cocktail) and trypsin. (B) Down-regulated viral proteins after A3 treatment (fold change >1.3), popularly reported proteins presented in pink color. (C) Inhibitory activity of A3 against nucleoprotein at 20 μM determined by enzyme linked immunosorbent assay (ELISA) kit (Abclonal, https://abclonal.com.cn/), and the positive control was PJ-34 [50]. (D) Four categories Q1–Q4 were divided by A3/mock ratio (<0.667, 0.667–0.769, 1.3–1.5 and > 1.5), and the numbers of their phosphoproteomic sites were presented. Mock group: Vero E6 cells infected by SARS-CoV-2 for 24 h; A3 group: Vero E6 cells infected by SARS-CoV-2 in the presence of A3 (10 μM) for 24 h, n = 3. (E) Enrichment analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways for categories Q1–Q4. (F) Mitogen-activated protein kinase (MAPK) signaling pathway analysis to explain the anti-inflammatory mechanism of A3. (G) Expression of proteins participated in inflammation-related MAPK signaling pathway in SARS-CoV-2 infected Vero E6 cells before and after the treatment of A3. Mock group: Vero E6 cells infected by SARS-CoV-2 for 24 h; A3 group: Vero E6 cells infected by SARS-CoV-2 in the presence of A3 (10 μM) for 24 h, n = 3. (H) Interleukin (IL)-1, tumor necrosis factor (TNF)-α, IL-6, IL-1β, IL-8, IL-7, function of interferons (IFN)-β and IFN-γ of proteomic samples were measured by monkey ELISA kit (MEIMIAN, www.mmbio.cn). Vero E6 cells respectively treated with SARS-CoV-2 in the absence (control), and presence of A3 for 24 h. ∗P < 0.05, ∗∗P < 0.01, compared with the control group, n = 3. LC-MS/MS: liquid chromatography coupled to tandem mass spectrometry; TMT: tandem mass tags.