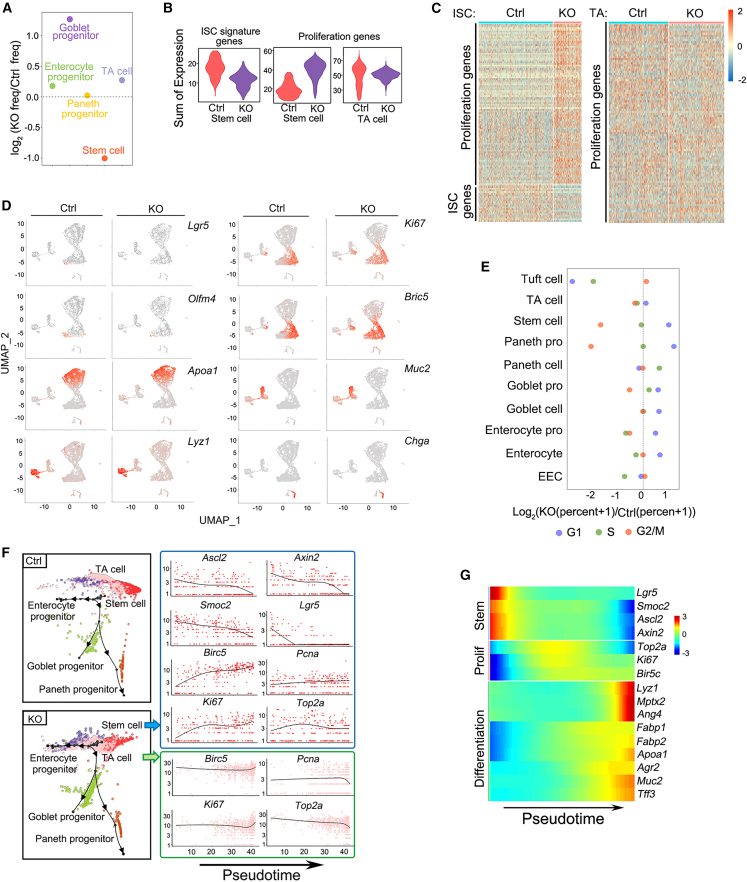
Figure 3.
Mettl3 deletion results in downregulation of the stem cell signature genes
(A) Dot plot depicting the relative frequency of stem cells, TA cells, and progenitor cells in control and Mettl3-KO-GFP mice at 4 dpt, revealed by scRNA-seq.
(B) Violin plots showing the metagene expression levels per single cell of the stem cell signature genes and proliferation genes in stem cells and TA cells of control and Mettl3-KO-GFP mice at 4 dpt revealed by scRNA-seq.
(C) Expression heatmap of the stem cell signature genes and proliferation genes in stem cells (left) and TA cells (right) in control and Mettl3-KO-GFP mice.
(D) Expression of the stem cell marker genes (Lgr5, Olfm4), the proliferation marker genes (Ki67, Birc5), and the differentiation cell marker genes (Apoa1, Lyz1, Muc2, Chga) in intestinal epithelium from control and Mettl3-KO-GFP mice were plotted by UMAP. Color from gray to red indicates relative expression levels from low to high.
(E) Dot plot depicts the relative percent of the cells in the G1, G2/M, and S phase in control and Mettl3-KO-GFP mice.
(F) Pseudotime analysis of stem cells, TA cells, and progenitor cells in control and Mettl3-KO-GFP mice at 4 dpt based on scRNA-seq (left). The expression of stem cell markers (Ascl2, Axin2, Smoc2, and Lgr5) and proliferation markers (Birc5, Top2a, Ki67, and Pcna) in stem cells and TA cells of Mettl3-KO-GFP mice was plotted along the pseudotime axis (right).
(G) Heatmap showing the transcription trends of marker genes of stem cells, TA cells, and differentiated cells along the pseudotime. Color scale represents Z score normalized expression levels. The pseudotime line contains all control and Mettl3-KO-GFP cells with the control cell cluster as the starting point. See also Figure S5.