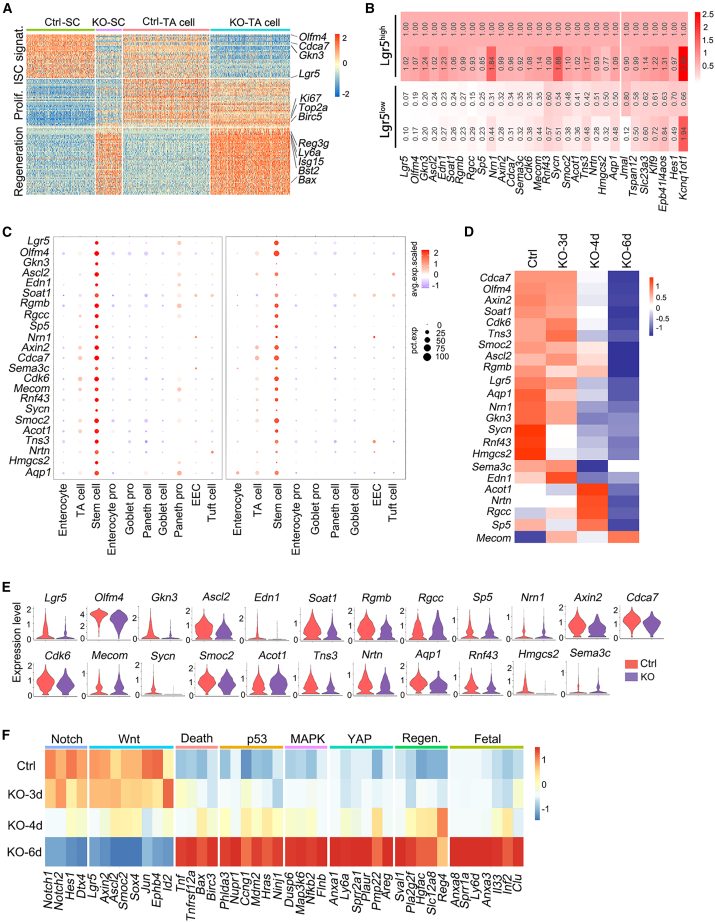
Figure 4.
Identification of the stemness genes via bulk RNA-seq and scRNA-seq analysis
(A) Heatmap of differentially expressed genes in stem cells and TA cells of control and Mettl3-KO-GFP mice at 4 dpt based on scRNA-seq.
(B) RNA-seq analysis of the relative expression of genes, which were enriched in Lgr5high cells in control mice.
(C) scRNA-seq analysis showing the gene expression level (color scale) and expressing cells (point diameter) in each cell cluster of the intestinal epithelium. All of these genes were sifted out of the gene list (B) showing specific expression in stem cells in our scRNA-seq data (left) and published ileum scRNA-seq data (right).
(D) Expression heatmap of the filtered 23 stemness genes in Lgr5+ ISCs of control and Mettl3-KO-GFP mice at 3, 4, and 6 dpt, based on bulk RNA-seq.
(E) Violin plots showing the expression of the 23 stemness genes in stem cells of control and Mettl3-KO-GFP mice.
(F) Expression heatmap of different pathway genes in Lgr5+ ISCs of control and Mettl3-KO-GFP mice at 3, 4, and 6 dpt, based on bulk RNA-seq. See also Figure S5.