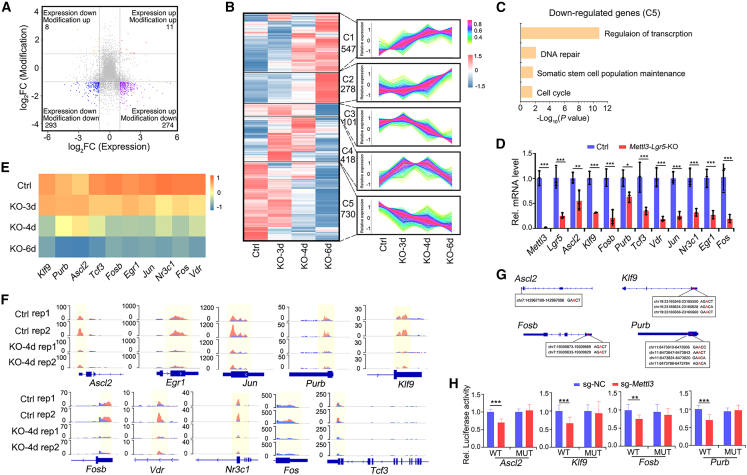
Figure 5.
Identification of transcriptional factors that promote stemness
(A) Distribution of the genes that exhibited different m6A modification and gene expression levels in control or Mettl3-KO-GFP Lgr5high cells (|log2FC|≥1; p < 0.05). The gene number in each category is shown.
(B) The expression of the genes with reduced m6A modification in Mettl3-KO-GFP Lgr5+ cells were divided into five modules according to their expression dynamics. The change trend in each module is shown in the line chart on the right.
(C) Functional enrichment analysis of the genes whose expression was downregulated at 3 dpt from the module C5 in (B).
(D) Expression of Mettl3, Lgr5, and the 10 TFs (Ascl2, Klf9, Fosb, Purb, Tcf3, Vdr, Jun, Nr3c1, Egr1, Fos) in FACS-sorted Lgr5high cells from control and Mettl3-Lgr5-KO mice at 4 dpt by q-PCR. N = 3 mice per group.
(E) Expression heatmap of the 10 TF candidates in Lgr5+ ISCs from control and Mettl3-KO-GFP mice, based on bulk RNA-seq.
(F) Integrative Genomics Viewer (IGV) tracks displaying MeRIP-seq reads along the indicated mRNAs in Lgr5high ISCs of control and Mettl3-KO-GFP mice. Blue reads are from input libraries and red reads from anti-m6A immunoprecipitation libraries. The y axis represents the CPM (count per million) of genes. The yellow boxes of the tracks depict the positions of m6A peaks.
(G) Representative m6A modification sites at the genomic loci of Ascl2, Klf9, Fosb, and Purb.
(H) Fetal human colon cells (FHC cells) were transfected with WT or MUT 3′ UTR luciferase reporter, as well as with control sgRNA or Mettl3 sgRNA, and luciferase activity was determined 48 h later. NC: control. MUT: mutation all sites (A to T). Data from three independent experiments were combined and are shown. The data represent mean ± SD. ∗∗∗p < 0.001, ∗∗p < 0.01, two-way ANOVA (D and H). See also Figure S6 and Tables S2–S4.