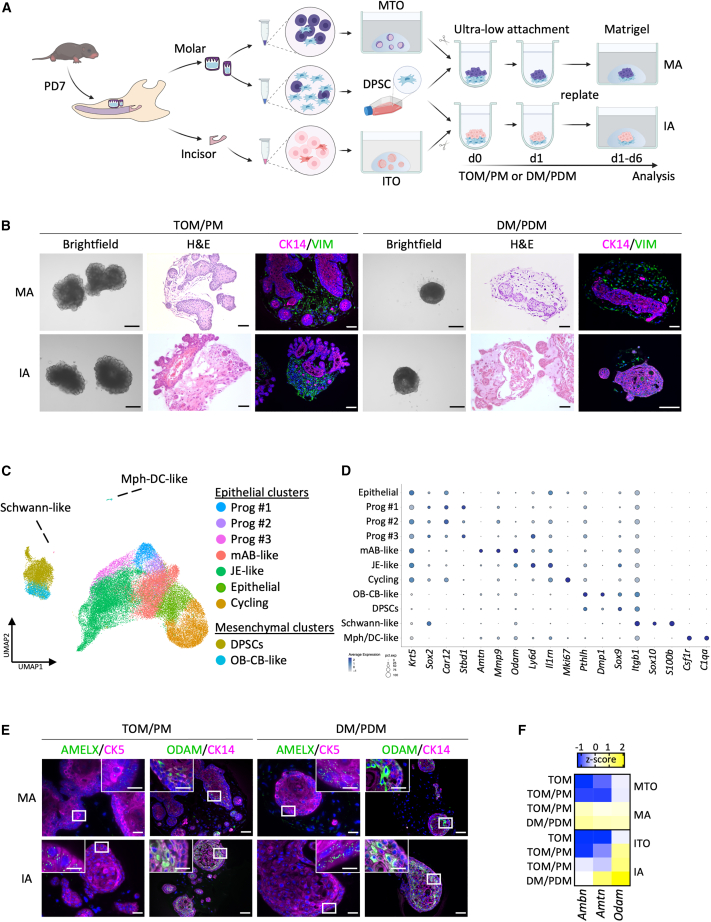
Figure 4.
Establishment and characterization of tooth assembloids combining organoid DESCs with mesenchymal DPSCs
(A) Schematic of experimental setup for the development of tooth assembloids by combining TO cells with mesenchymal DPSCs.
(B) Bright-field, H&E, and IF (of indicated markers) images of MAs and IAs grown in TOM/PM or DM/PDM. Nuclei of IF images are counterstained with Hoechst33342.
(C) Annotated UMAP plot of integrated assembloid scRNA-seq datasets, i.e., from MA+TOM/PM, MA+DM/PDM, IA+TOM/PM, and IA+DM/PDM (n = 2 biological replicates of each experimental condition).
(D) Dotplot displaying the percentage of cells (dot size) expressing key marker genes of the different annotated cell clusters (average expression levels indicated by color intensity; see scale).
(E) IF analysis of AMELX and ODAM (green) and indicated cytokeratins (CK; magenta) in tooth assembloids. Nuclei are counterstained with Hoechst33342 (blue). Boxed areas are magnified.
(F) Heatmap of gene expression of AB-resembling differentiation in tooth assembloids as quantified by qRT-PCR analysis. Data are presented as relative expression to Gapdh (ΔCt) and Z score normalized. Colors range from blue (low expression) to yellow (high expression).
Scale bars: 250 μm for bright-field images, 50 μm for H&E, CK14/VIM, and ODAM/CK14 (20 μm for close-ups) IF images, 25 μm for AMELX/CK5 (10 μm for close-ups) IF images. See also Figure S4.