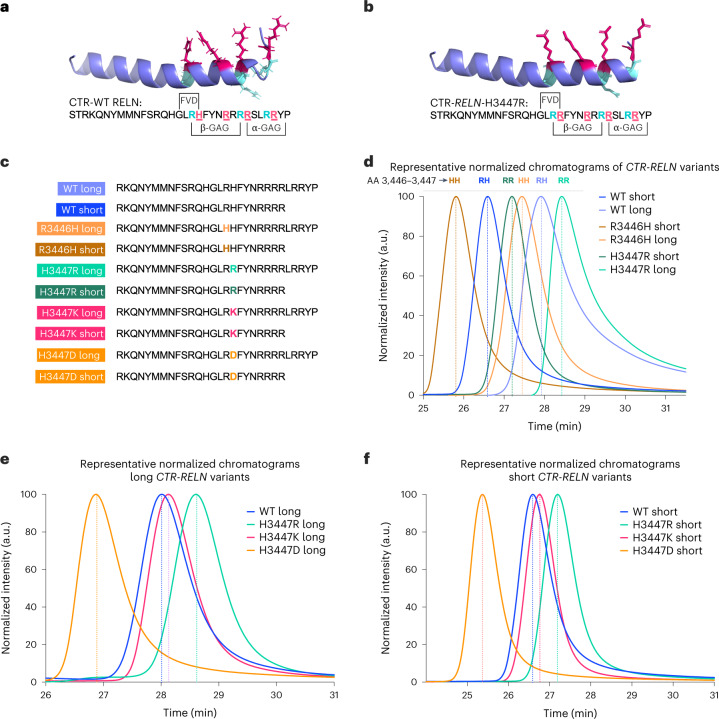
Fig. 3. Binding profiles of CTR-RELN variants with heparin.
a,b, Representative in silico models depicting the orientation of basic amino acids in the heparin-binding motif (highlighted with colors) using the WT CTR-RELN NMR structure (a). For the H3447R CTR-RELN variant (b) the model was determined by a homology-based model of WT CTR-RELN that was calculated by Swiss-Model. Position 3,447 orients in the same direction as most other arginines (magenta). Arginines in positions 3,446, 3,453 and 3,457 (cyan) may also interact with heparin as part of the heparin-binding motif but are oriented differently from most basic amino acids. c, RELN peptide variant sequences used for the HPLC analysis. d, Representative chromatographic profiles normalized to the maximum of each eluted peak of short or long CTR-RELN peptides with zero (R3446H, orange), one (WT, blue) or two (H3447R, green) arginines in positions 3,446–3,447, which are predicted to contribute to increased interaction with heparin and are indicated by later peak retention time in the isocratic 1 M KCl elution. e,f, Long (e) or short (f) CTR-RELN peptides with zero (R3446H, orange), one (WT, blue) or two (H3447R, green) arginines in positions 3,446–3,447 showing increased interaction with heparin for the long variants, as indicated by later peak retention time in the isocratic 1 M KCl elution. Conversely, short RELN variants have earlier peak retention times compared to long RELN variants (e). n = 2 independent chromatographic repeats within <0.5 min of representative peaks. Data are expressed as normalized to the maximum emission wavelength for each peak.