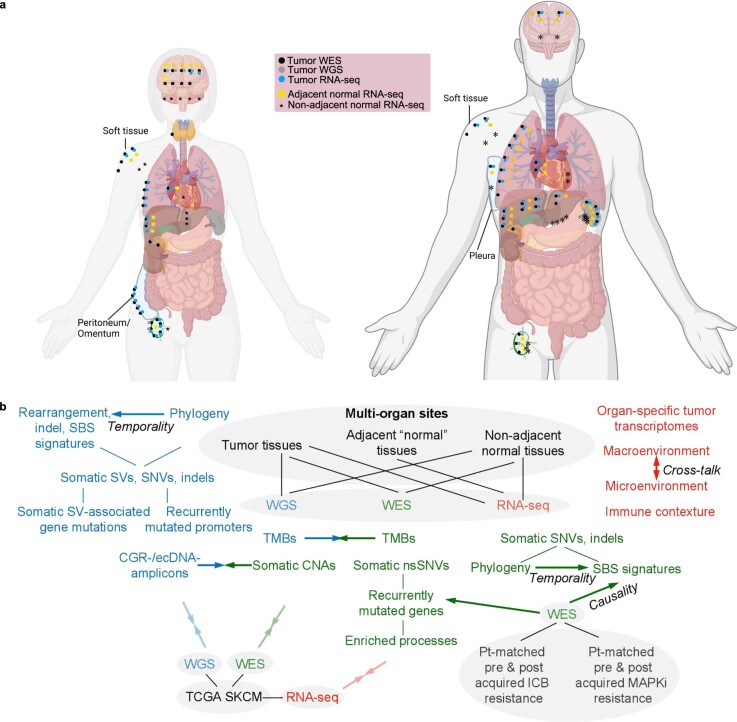
Extended Data Fig. 1. Overview of the tissues sampled at rapid autopsies and the study design.
(a) Numbers and types of tissues, sequencing strategies, and metastatic sites are shown for 11 rapid melanoma autopsy (RAM) cases consisting of five deceased female patients and six deceased male patients with BRAFMUT (n = 7) or NRASMUT (n = 4) cutaneous melanoma. Not represented are RNA-seq derived from triplicates and WES derived from non-adjacent normal tissues. (b) Inclusion of tissue sets and datasets (grey background) and flow chart of analyses performed. RAM is the core tissue or data set, supplemented by comparisons (transparent, adjoined arrows pointing in opposite directions) with two tissue/data sets. RAM cohort (tumor, n = 22 for WGS, n = 74 for WES, n = 93 for RNA-seq; adjacent ‘normal’, n = 68 for RNA-seq; non-adjacent normal, n = 67 for RNA-seq, n = 10 for WES). Pre-and-post cohort consists of patient-matched cutaneous melanoma tumors biopsied pretreatment and post acquired resistance (ICB, n = 7 pairs; MAPKi, n = 59 pairs; both subgroups with patient-matched normal tissues for all patients; n = 102 tumor WES datasets). TCGA–SKCM cohort, only BRAFMUT (n = 233) or NRASMUT (n = 125) tumors included (13 tumors with both BRAF and NRAS mutations; total n = 345 tumors). Bolded arrows, analyses compared to derive insights into temporality or causality.