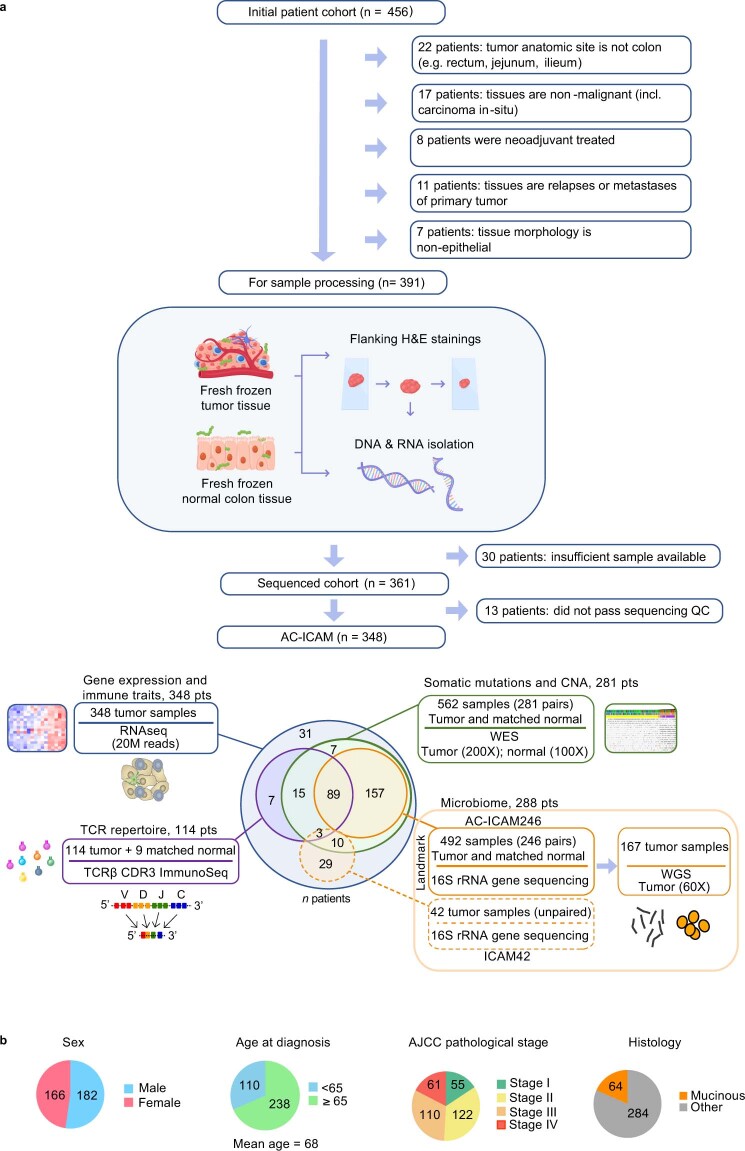
Extended Data Fig. 1. Study design for comprehensive genomic profiling of colon cancer.
AC-ICAM study design. a, Visual representation of exclusion criteria and number of excluded samples from the 456 available samples in the LUMC biobank, followed by overview of tissue processing and genomic profiling of fresh-frozen tumor and matched normal colon tissue samples. Samples of a total of 348 colon cancer patients were included in AC-ICAM. Number of profiled samples and technical specifications are indicated for each platform, including RNA Sequencing (RNA-Seq), Whole-Exome Sequencing (WES), TCR sequencing (immunoSEQ TCRβ assay) and 16 S rRNA gene sequencing to profile the microbiome. AC-ICAM246 is a subset of AC -ICAM with tumor–normal matched rRNA 16 S microbiome data, while AC-ICAM42 only has tumor samples with 16 S rRNA gene sequencing. Venn diagram reflects overlap in number of patients between the different platforms applied. b, Summary of patient characteristics of colon cancer cohort (n = 348). Number in pie chart indicates number of patients in each category.