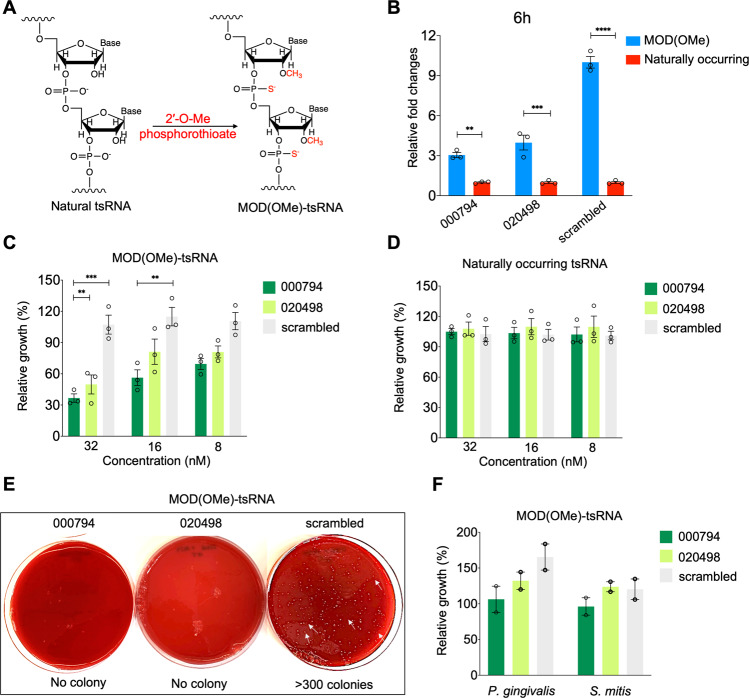
Fig. 2. Chemically modified Fn-targeting tsRNAs confer superior growth inhibition of Fn in a sequence-, and species-dependent manner.
A Incorporation of 2’-OMe phosphorothioate linkage in tsRNAs, referred to as MOD(OMe)-000794, MOD(OMe)-020498 and MOD(OMe)-scrambled RNA. B Improvement of tsRNA stability by chemical modifications in comparison to synthetic mimics of naturally occurring counterparts. Individual tsRNAs (1 nM) were incubated with Columbia broth anaerobically at 37 °C for 6 h, and intact tsRNAs were measured via quantitative real time PCR using a stem-loop method. Fold changes were normalized to the levels of naturally occurring tsRNAs, which are shown as “1” on the y axis. n = 3 technical replicates, N = 2 independent experiments. C Growth inhibition of Fn ATCC 23726 by MOD(OMe)-000794 and MOD(OMe)-020498, but not MOD(OMe)-scrambled RNA, at the nanomolar concentration ranges (n = 3, N = 3). D Naturally occurring tsRNAs failed to inhibit Fn ATCC 23726 at indicated concentrations (n = 3, N = 3). E Representative images showing the colony formation of Fn ATCC 23726 on agar plates after treatment with 512 nM MOD(OMe)-000794, MOD(OMe)-020498 or MOD(OMe)-scrambled RNA for 24 h. Bacteria were washed and recovered on nonselective Columbia broth sheep blood agar plates. Representative colonies are indicated by white arrows. F MOD(OMe)-000794 and MOD(OMe)-020498 at 512 nM exhibited no growth inhibition in two representative oral bacteria, a Gram-negative bacterium, Porphyromonas gingivalis ATCC 33277 (P. gingivalis) and a Gram-positive bacterium, Streptococcus mitis ATCC 6249 (S. mitis). (n = 2, N = 2). Data were analyzed by the two-way ANOVA followed by Dunnett’s Bonferroni multiple comparison tests. **p < 0.01, ***p < 0.001, ****p < 0.0001. Data are means ± SEM.