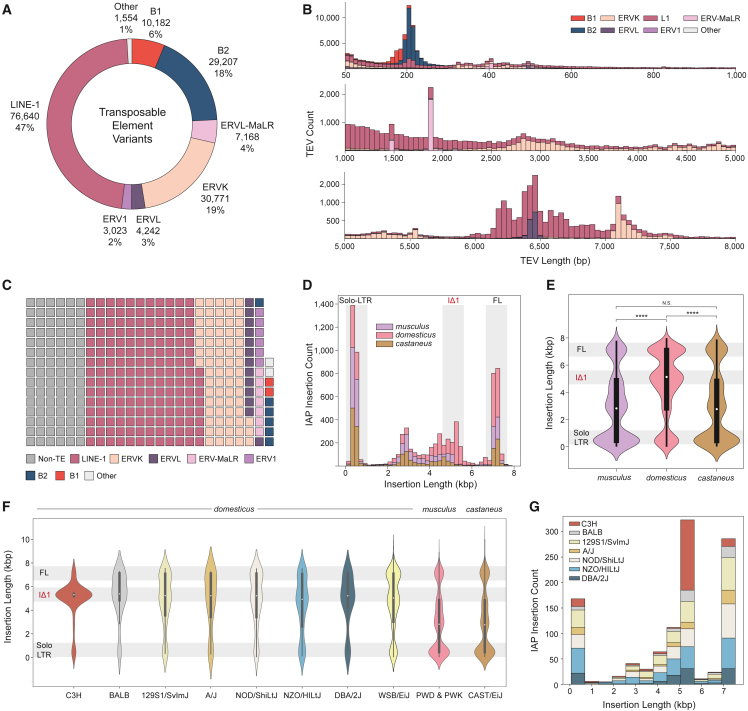
Figure 2.
TEV in diverse mouse genomes
(A) Total number of TEVs discovered from long read sequencing by transposon family.
(B) Length distribution of TEVs by transposon family.
(C) Total number of variable bases due to TEV by transposon families and non-TE SVs. Each block equals one megabase of variable sequence.
(D) Count of subspecies-specific intracisternal A-particle (IAP) variants. Highlighted size ranges are solo-LTR (∼300 bp), IΔ1 variants (4.5–5.5 kbp), and full-length (FL) variants (6.5–7.5 kbp).
(E) Size distribution of species-specific IAP insertions (Mann-Whitney U test; ∗∗∗∗p ≤ 1 × 10−4).
(F) Size distribution of strain-specific IAP insertions within the domesticus lineage. Substrains that share a parental origin (C3H, C3H/HeJ and C3H/HeOuJ; BALB, BALB/cByJ and BALB/cJ) are grouped to represent each lineage.
(G) Count of strain-specific IAP insertions in closely related domesticus animals.