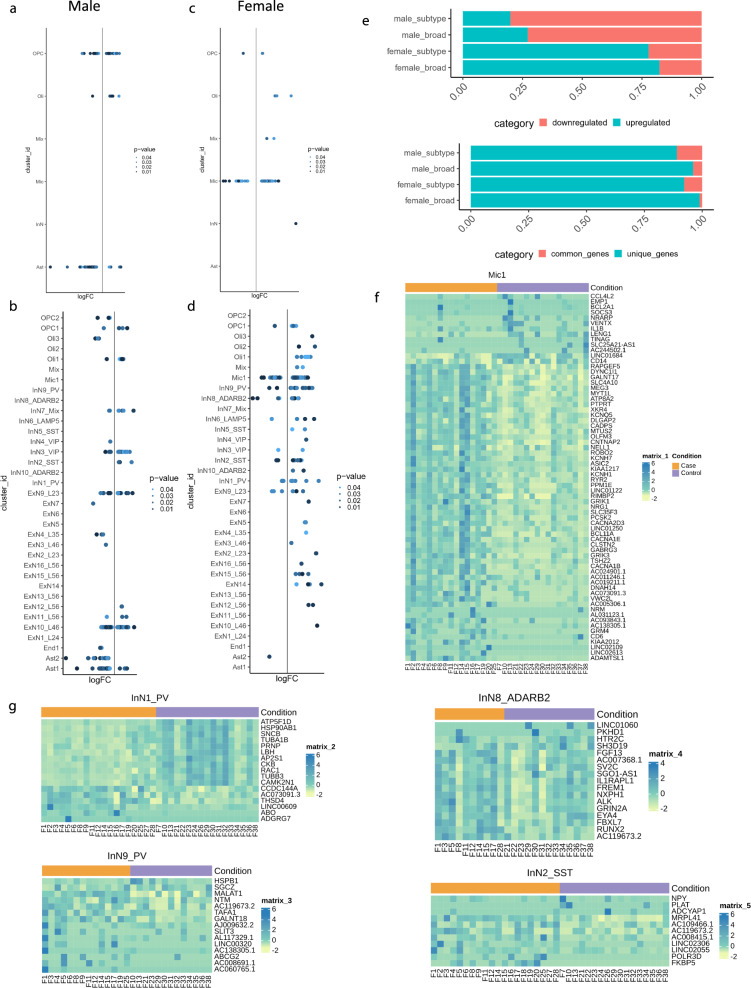
Fig. 3. Cell type specific differential gene expression in males and females with MDD.
a, b Distribution of differentially expressed genes in (a) broad cell types and (b) clusters with increased and decreased expression in male cases compared to controls. c, d Distribution of differentially expressed genes in (c) broad cell types and (d) clusters with increased and decreased expression in female cases compared to controls. For a–d, points are colored by the corrected p-value for differential expression, and upregulated genes are plotted to the right of the midline while downregulated genes are plotted to the left. e Barplots showing proportions of up and downregulated genes and unique and shared genes. For males, the majority of DEGs were decreased in expression in cases compared to controls both at the broad (110/151, 73%) and cluster levels (358/447, 80%) and most DEGs were cell type specific both at the broad (145/151, 96% unique DEGs) and cluster (398/447, 89% unique DEGs) level. For females, the majority of DEGs were upregulated both at the broad (70/85, 82%) and the cluster level (140/180, 78%) and most DEGs were cell type specific both at the broad (84/85, 99% unique DEGs) and cluster (166/180, 92% unique DEGs) level. f, g Heatmaps showing the pseudobulk expression of differentially expressed genes in top clusters with highest number of DEGs in the female cluster level analysis—f microglia, g inhibitory neuronal clusters. For f, g, the plotted values are pseudobulk CPMs (counts per million) calculated with edgeR and muscat and scaled per row (by gene). For all heatmaps (f, g), the annotation bar at the top is colored orange for cases and purple for controls, and rows and columns are not clustered. Statistical testing corresponding to Fig. 3a–d were performed with the edgeR (glmQLFit, glmQLFtest), FDR (Benjamini & Hochberg) corrected p-values are plotted. Source data are provided as a Source Data file.