Figure 3.
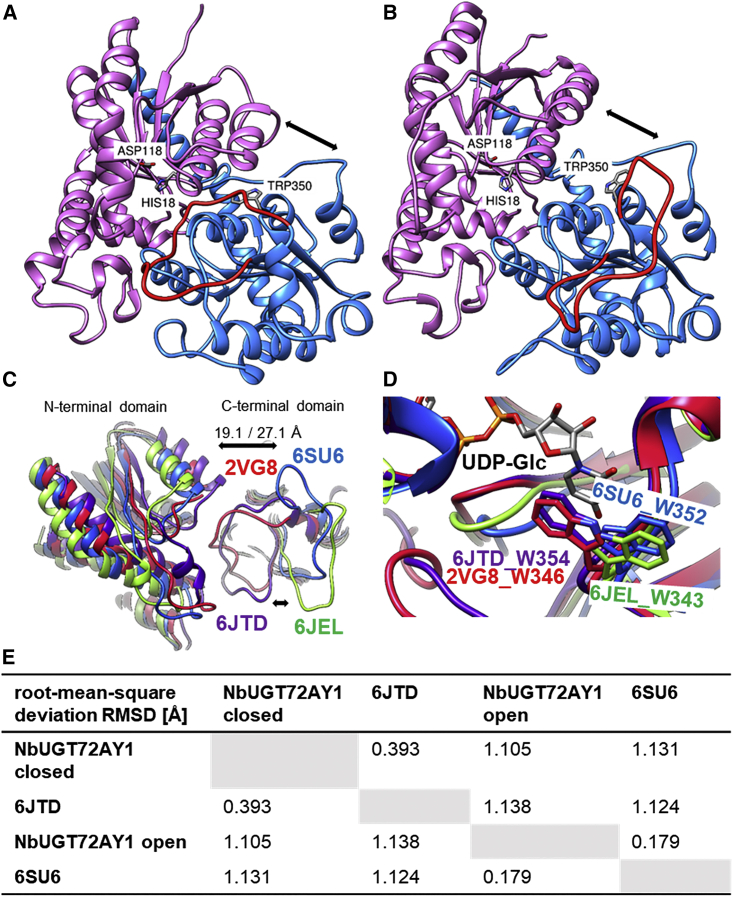
3D UGT structures showing closed and open conformers.
(A) Prediction of the 3D structure of NbUGT72AY1 (closed conformer) was performed by the IntFOLD Integrated Protein Structure and Function Prediction Server (https://www.reading.ac.uk/bioinf/IntFOLD/) with default values based on 6JTD (www.rcsb.org). The result was visualized with UCSF Chimera (https://www.cgl.ucsf.edu/chimera). N- and C-terminal domains are shown in purple and blue, respectively. Important amino acids are marked, and the flexible loop covering the catalytic site is highlighted in red.
(B) Prediction of the 3D structure of NbUGT72AY1 (open conformer) was performed by the SWISS-MODEL Server (https://swissmodel.expasy.org) with default values based on 6SU6 (www.rcsb.org).
(C) Close-up of the superimposition of two putative open UGT conformers (6SU6 and 6JEL) and two closed UGT conformers (6JTD and 2VG8). Distances were measured between the N- and C-terminal domains of 6JTD and 6SU6 (S62.A CB to E263.A CB in 6JTD: 19.1 Å, in comparison to S62.A CB to E262.A CB in 6SU6: 27.1 Å).
(D) Tryptophan/uridine π-stacking interaction of the first amino acid of the PSPG box and UDP-Glc. In the crystal structures of the putative open protein conformers (6SU6 and 6JEL), W is rotated by 180° in comparison to the 3D structures of the closed conformers (6JTD and 2VG8).
(E) Calculation of mutual root-mean-square deviation (RMSD) values using UCSF Chimera.