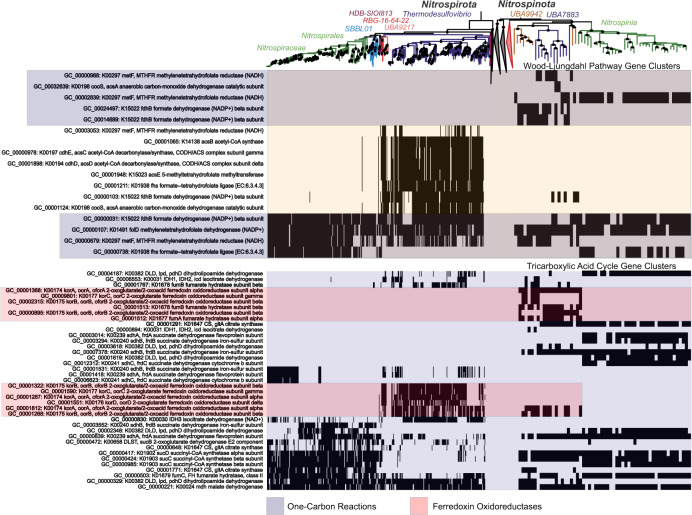
Fig. 5. Presence/absence patterns of gene clusters with annotations involved in the Wood-Ljungdahl Pathway (WLP, KO Ids included in KEGG Module M00377) and tricarboxylic acid cycle (TCA, KO Ids included in KEGG Modules M00009-11) in Nitrospirota and Nitrospinota.
The rows are ordered by hierarchical clustering of presence/absence patterns in Nitrospirota using Ward’s Linkage. Functional differences and similarities between the two phyla can be noted, namely the use of WLP in early-branching Nitrospirota and the shared use of the rTCA cycle in Nitrospirales and Nitrospinota. The purpled shaded boxes denote gene clusters with annotations involving C1 metabolisms, the red shaded boxes denote kor/oor annotated gene clusters likely involved in low redox-potential ferredoxin cycling present in basal groups of both phyla.