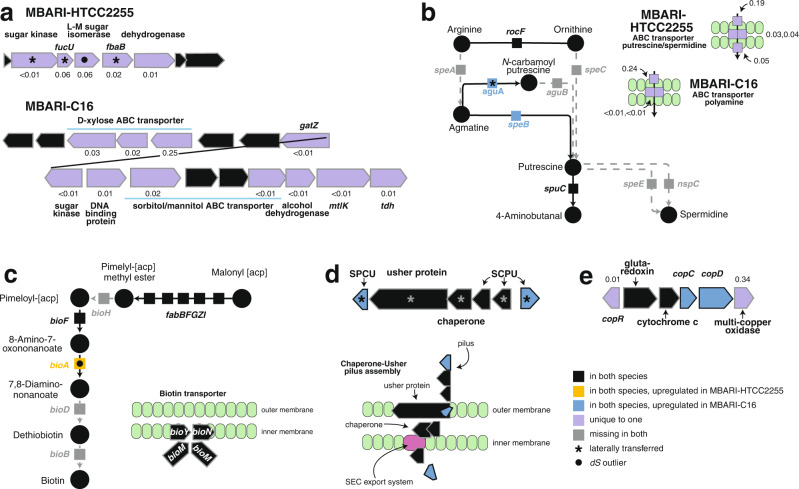
Fig. 4. Gene composition and expression divergence between MBARI-HTCC2255 and MBARI-C16.
a Sugar uptake and catabolism genes. fucU, L-fucose mutarotase; L-M, lyxose-mannose; fbA, fructose-bisphosphate aldolase; gatZ, tagatose-1,6-bisphosphate aldolase; mtlK, mannitol-2-dehydrogenase; tdh, threonine dehydrogenase. b Polyamine synthesis and uptake genes. speA, arginine carboxylase; speB, agmatinase; speC, ornithine decarboxylase; speE, spermidine synthase; aguA, agmatine deiminase; aguB, N-carbamoylputrescine amidase; spuC, putrescine-pyruvate transaminase; nspC, carboxynorspermidine decarboxylase. c Biotin synthesis and uptake genes. bioA, adenosylmethionine-8-amino-7-oxononanoate aminotransferase. d Chaperone-usher (CU) pilus synthesis genes. SCPU, spore coat U domain-containing protein. e Copper-related operon. copR, transcriptional regulator; copC, copper binding protein; copD, copper resistance protein. Expression levels are shown adjacent to each unique gene in units of transcripts per gene copy; the average expression level across all genes and all sample dates is 0.07 transcripts per gene copy.