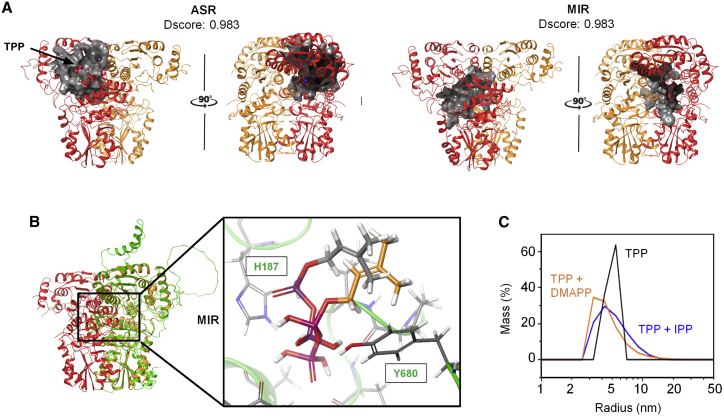
Figure 2.
Docking and DLS analysis of DXS–ligand binding.
(A) Docking simulation of the ligand binding site for EcDXS. The two predicted binding sites for ligands with high Sitemap scores (Dscores) are shown in gray: the active site region (ASR) and monomer interaction region (MIR).
(B) Structural superposition of EcDXS and SlDXS1. EcDXS (PDB: 2O1S) is marked with orange-red structures, and SlDXS1 (Alpha-Fold2 model AF-Q9XH50-F1) is represented in green. Magnification shows the MIR binding mode prediction from docking calculations of DMAPP for EcDXS (gray carbon atoms) and SlDXS1 (orange carbon atoms). SlDXS1 residues interacting with DMAPP are indicated.
(C) DLS analysis of purified SlDXS1 with the indicated ligands. Recombinant SlDXS1 (20 μM) was pre-mixed with 50 μM TPP and analyzed in the absence (black line) or presence of 50 μM IPP (blue line) or 50 μM DMAPP (orange line). For each measurement, 5 acquisitions of 5 s were taken.