FIGURE 2.
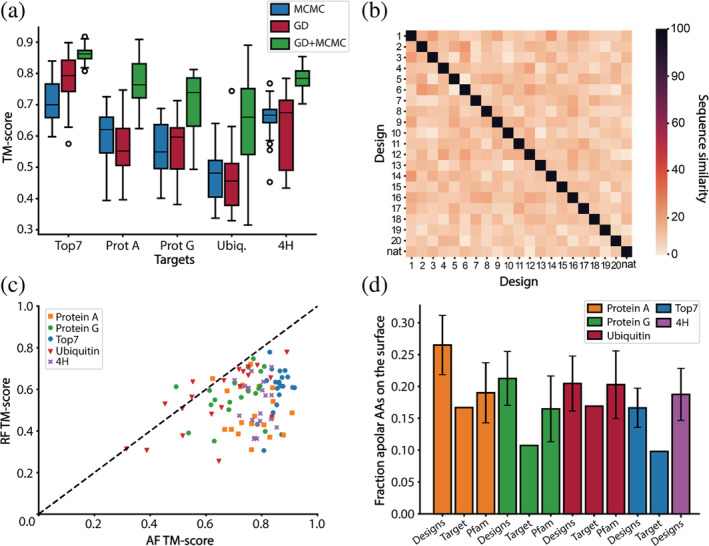
Features of AlphaFold2 (AF2) designed sequences. (a) TM‐scores of designed sequences using Markov Chain Monte Carlo (MCMC), gradient descent (GD) optimization and a combination of both (GD + MCMC). Blue–TM‐scores after MCMC design; red–TM‐scores after GD design; green–TM‐scores after GD design followed by MCMC optimization. For each fold, 20 rounds of GD and MCMC optimization were performed. A combination of GD and MCMC optimization significantly improves the TM scores compared to MCMC and GD only designs. (b) Evaluation of the sequence diversity obtained within the top7 designs. The designed sequences have a low sequence similarity (between 10% and 30%) when compared to one another and to the native sequence. (c) Structure prediction of the AF2 designed sequences using AF2 and RF. In most instances, RF predicts lower TM‐scores than AF2. (d) Fraction of hydrophobics on the surface before the surface redesign step. All the designs have more hydrophobic residues on their surface than their target fold. When comparing the designs to their protein family, we find that the designs of protein A and protein G have slightly more hydrophobics on their surface than similar folds found in nature. Top7 and 4H are de novo proteins hence do not have a protein family, additionally, 4H is a backbone model designed with the TopoBuilder and as such there is no sequence to be compared to.
