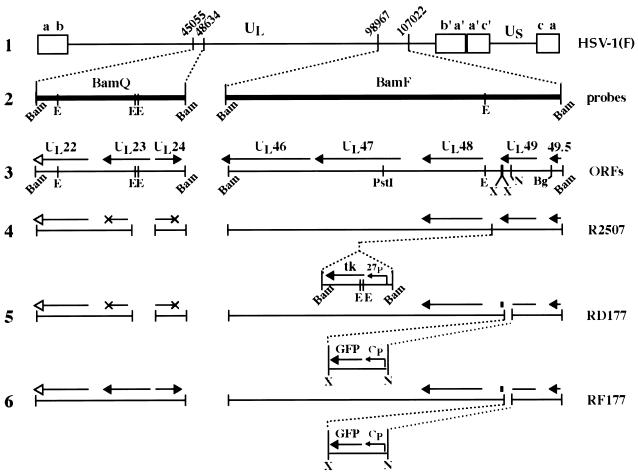
FIG. 1.
Schematic representations of the HSV-1 genome and viruses used in this study. The wild-type HSV-1 genome is shown in line 1. The unique long (UL), unique short (Us), and terminal repeat segments (a, b, c, a′, b′, and c′) are indicated. The coordinates of the BamQ and BamF fragments in the viral genome are indicated, and these regions are expanded in line 2. Line 3 shows the predicted open reading frames in the BamQ and BamF regions of wild-type virus. Solid arrowheads mark translation stop sites and show the direction of transcription; arrow length indicates approximate transcript size. The UL22 transcript designated with an open arrow possesses a translational stop site beyond the boundaries of the BamQ fragment. R2507 (line 4) is a derivative of the parental strain Δ305. Thus, R2507 possesses a deletion in the BamQ fragment which removes 500 bp, including a portion of the UL23 gene encoding the viral tk and a portion of the UL24 gene. These disrupted transcripts are denoted by an X at the locations of their stop sites in the wild-type genome. In R2507, a 1.8-kb tk expression cassette containing the viral tk gene under control of the ICP27 promoter (27p) has been inserted approximately 200 bp upstream of the UL48 start site (46). RD177 (line 5) and RF177 (line 6) were generated for the present study as described in Materials and Methods. Both viruses express the GFP under control of a cytomegalovirus promoter (Cp). The small solid rectangles in lines 5 and 6 indicate that the GFP cassette was inserted 45 bp 5′ of the UL49 stop site. The deletion in the BamQ locus present in R2507 and RD177 was repaired in RF177. Restriction sites: Bam, BamHI; E, EcoRV; X, XhoI; N, NsiI; Bg, BglII.