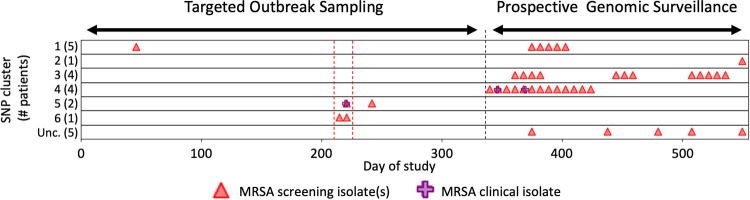
FIG 1.
NICU outbreak SNP cluster occurrence. x axis indicates day during the analysis period. “Targeted Outbreak Sampling” refers to microbiologic-only assessments of NICU strains from days 1 to 341. Triangles and crosses indicate all strains among patients who were also included in later genomic analyses. “Prospective Genomic Surveillance” over days 342 to 558 indicates the period of prospective genomic analyses of new strains. y axis indicates individual NCBI Pathogen Detection SNP clusters; numbers in parentheses indicate number of NICU strains from this study that occurred in the broader NCBI cluster, as follows: SNP clusters (MLST, SCCmec type): 1, PDS000067849.1 (8, IVa[2B]); 2, PDS000069849.1 (72, IVa[2B]); 3, PDS000069850.1 (5, II[2A]); 4, PDS000069851.1 (5, IVa[2B]); 5, PDS000069882.1 (1535, unclassified SCCmec); 6, PDS000069883.1 (8, IVa[2B]); Unc., unclustered isolates with unrelated MLST and SCCmec types. Red dashed lines show a suspected outbreak during targeted outbreak sampling, and a black dashed line marks when prospective surveillance began.