FIG. 1.
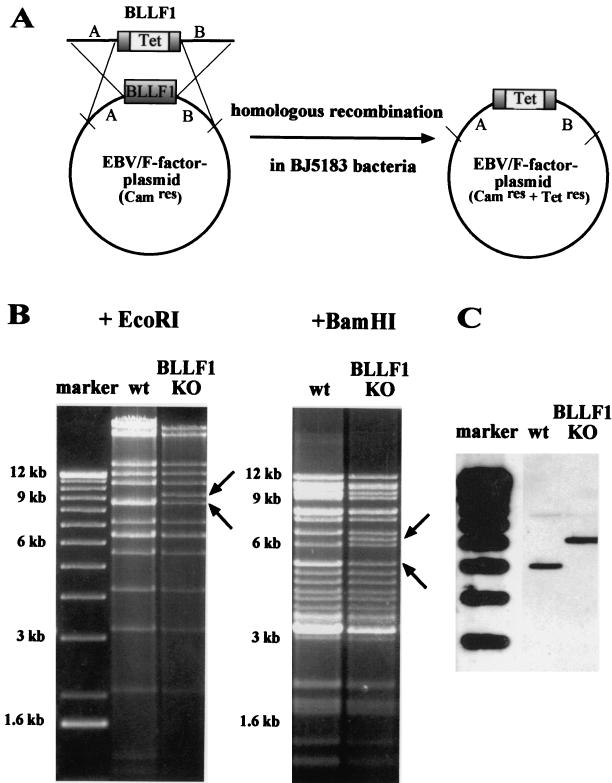
Mutant EBV lacking its major glycoprotein BLLF1. (A) Construction of the BLLF1-negative EBV mutant genome by homologous recombination in the recBCD E. coli strain BJ5183. The diagram shows schematically the wild-type EBV genome carrying the BLLF1 gene and a linearized fragment in which most of the BLLF1 sequence was replaced by the gene for tetracycline resistance (Tet). Since this linearized fragment used for targeted allelic exchange also carries EBV sequences that flank the BLLF1 gene, homologous recombination with the wild-type EBV F factor plasmid can take place. After transformation of the BLLF1/Tet fragment into the bacterial strain BJ5183, clones carrying recombinant EBV plasmids were selected for chloramphenicol (Cam) and tetracycline resistance. (B) Restriction fragment analysis of EBV BLLF1-KO DNA in comparison with wild-type B95.8/F factor DNA (wt). BLLF1-negative EBV plasmid DNA was purified from a chloramphenicol- and tetracycline-resistant DH10B E. coli clone and digested with EcoRI or BamHI. The restriction pattern of BLLF1-KO mutant DNA was compared with the one obtained for wild-type EBV DNA. Modified DNA fragments are indicated by arrows. As expected from the predicted BamHI restriction pattern, the introduction of the tetracycline resistance gene into the BLLF1 gene leads to the appearance of an additional 6.2-kb BamHI fragment and the disappearance of one of the two 5-kb fragments that are seen after digestion of wild-type EBV DNA. Similarly, the EcoRI restriction pattern obtained after digestion of the BLLF1 mutant EBV DNA shows a predicted additional fragment of 9.3 kb that correlates with the disappearance of one of the two 8.5-kb fragments obtained when wild-type EBV DNA is analyzed. (C) Southern blot analysis of 293-BLLF1-KO cells harboring the BLLF1-negative EBV mutant strain compared to 293-B95.8/F cells carrying the wild-type EBV plasmid. Total genomic DNA from these cells was digested with the BamHI restriction enzyme. After agarose gel electrophoresis and Southern blotting, the DNA was hybridized with a probe derived from the BLLF1/Tet fragment that was used to introduce the BLLF1 KO mutation. The probe spanning nucleotide coordinates 88335 to 93167 of the B95.8 EBV sequence with a deletion of 90263 to 91916 (see Materials and Methods) identifies a characteristic 6.2-kb signal in DNA from 293-BLLF1-KO cells, compared to a 5-kb band observed in 293 cells carrying wild-type virus (see also panel B). The faint signals at 7.9 kb that are observed with BLLF1-negative virus and with wild-type virus represent remaining BLLF1 sequences that partially hybridize with the probe.