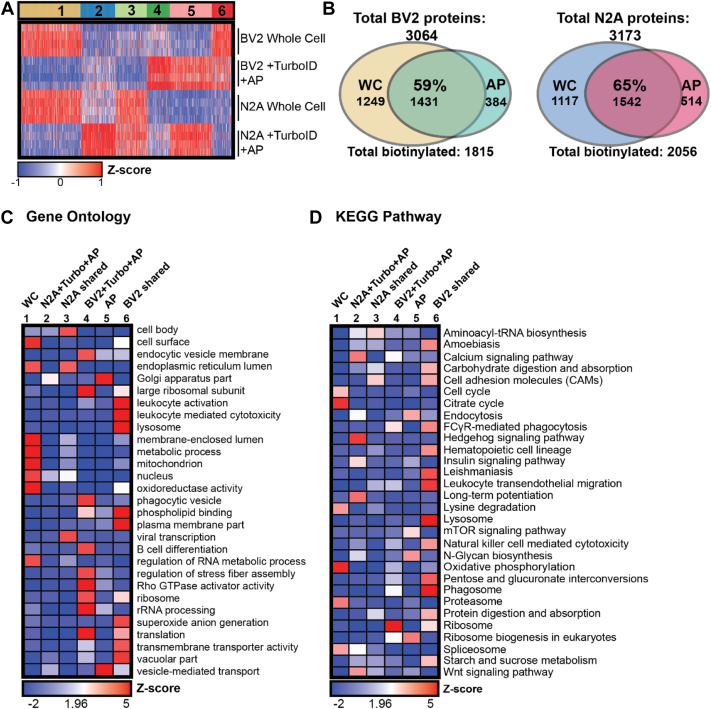
Fig. 3.
Global profiling of TurboID labeling in microglial and neuronal cell lines. A, K-means clustered heatmap representation of label-free quantitation (LFQ) intensity data of proteins identified by mass spectrometry (MS) in whole cell (WC) and biotinylated and affinity-purified (AP) proteomes (n = 4 per experimental group) in microglial and neuronal cell lines. Six distinct clusters represent labeling profile of TurboID, including proteomes enriched in WC preparations, AP preparations, and by cell type. B, Venn diagram of protein counts identified in BV2 WC samples and BV2 + TurboID + AP samples and N2A WC and N2A + TurboID + AP samples. In BV2 cells, TurboID-NES labels ∼59% of the total proteins captured by LFQ MS. In N2A cells, TurboID-NES captures ∼65% of the total proteins identified by LFQ MS. C, functional annotation of gene set enrichment based on Gene Ontology (GO) over-representation analysis (ORA). Heatmap color intensity is based on Z-score enrichment across the six clusters. D, heatmap representation and functional annotation of clusters derived from the KEGG pathways. KEGG, Kyoto Encyclopedia of Genes and Genomes; NES, nuclear export sequence.