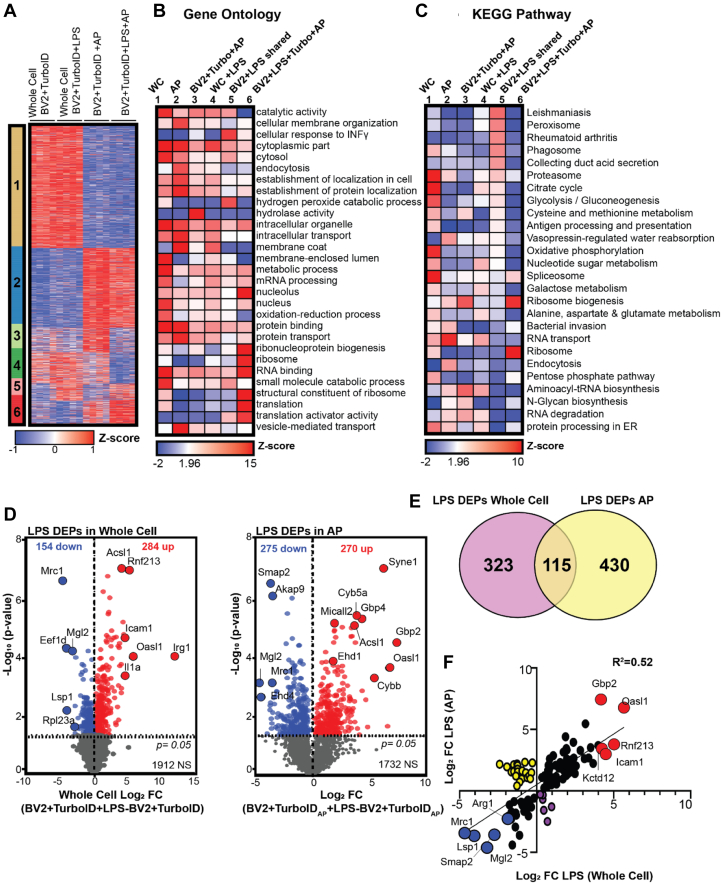
Fig. 6.
TurboID-mediated biotinylation partially captures lipopolysaccharide (LPS)-driven proteomic changes. A, K-means clustered heatmap representation of label-free quantitation (LFQ) intensity data of proteins differentially expressed by TurboID in whole-cell (WC) and transduced affinity-purified (AP) BV2 proteomes (n = 4 per experimental group). B, heatmap representation and functional annotation of Gene Ontology (GO) from murine GO database. Heatmap color intensity depicts Z-score values. C, heatmap representation of Z-scores associated with LPS differently expressed proteins (DEPs) derived from Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways database. D, differential expression analysis (DEA) of LPS DEPs in WC (left) and affinity purification (AP) (right) identifies 438 and 545 LPS DEPs, respectively. E, Venn diagram depicting 323 LPS DEPs unique to WC BV2 samples, 115 shared LPS DEPs, and 430 unique LPS DEPs in BV2 AP samples. F, linear regression of Log2FC values of BV2 AP samples (y-axis) and BV2 WC samples (x-axis) reveals a modest correlation (R2 = 0.52) of log2FC of the shared 115 LPS DEPs. Red points depict proteins the top five proteins significantly upregulated with LPS treatment in AP and WC samples. Yellow points represent proteins significantly increased in AP samples and significantly decreased in WC samples with LPS treatment. Blue points illustrate the top five proteins significantly decreased with LPS in both AP and WC BV2 samples. Purple points depict proteins that are significantly increased in the WC proteome and significantly decreased in the AP proteome with LPS treatment.