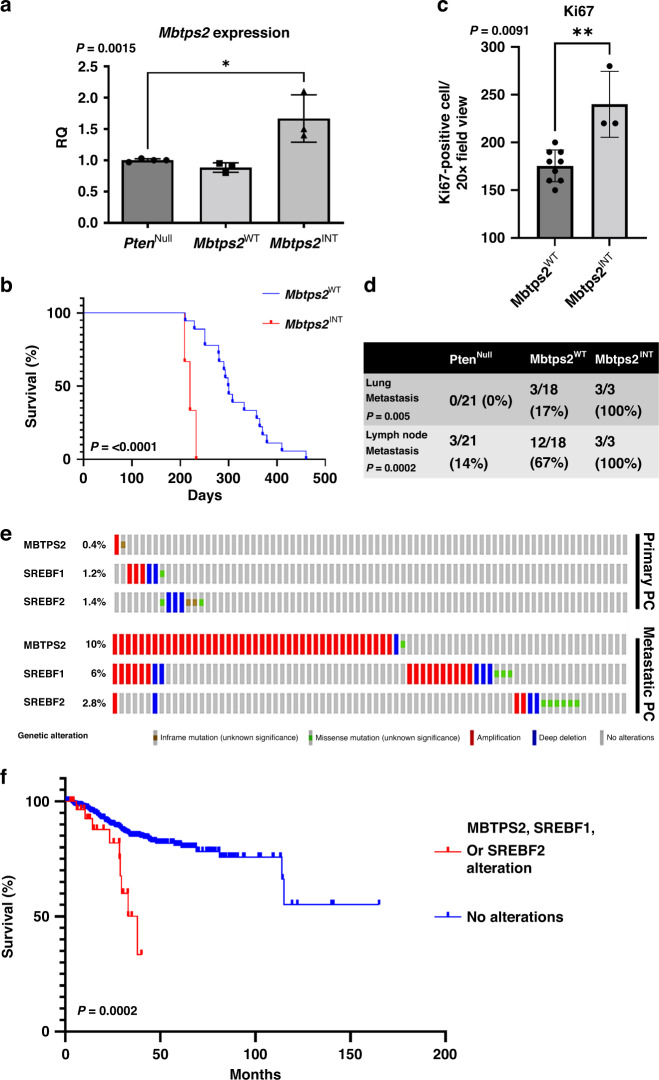
Fig. 2. Identification of MBTPS2 by Sleeping Beauty transposon screen on a background of PtenNull-driven prostate cancer.
a RT-PCR for Mbtps1 expression in prostate tumours from PtenNull, Mbpts2WT and Mbpts2INT mice (n = 4 mice for PtenNull, n = 3 mice for Mbtps2WT and Mbtps2INT, *p < 0.016; unpaired t-test). b Kaplan–Meier (log-rank) curve demonstrating reduced survival in the Mbtps2INT (n = 3) mice in comparison to Mbtps2WT (n = 18) mice, P < 0.0001; log-rank (Mantel–Cox) test. c Quantification of Ki67-positive cells in Mbtps2INT prostate tumours (n = 3 mice, minimum cell count of 100 cells per mouse) demonstrated increased levels of proliferation compared with tissue from Mbtps2WT tumours (n = 9, minimum cell count of 100 cells per mouse), **p = 0.0091; Mann–Whitney). d Percentages of mice with lung and lymph node metastases in Mbtps2INT mice compared to Mbtps2WT mice tested by Fisher’s exact test. e MBTPS2, SREBF1 and SREBF2 alterations visualised on CBioPortal using TCGA Firehose Legacy cohort for primary PC, and SU2C/PCF Dream Team(31) for the metastatic PC cohort. f Kaplan–Meier (log-rank) curve using TCGA Firehose Legacy cohort for primary PC, and SU2C/PCF Dream Team(31) showing patients with MBTPS2, SREBF1 or SREBF2 alterations (N = 31) and without these alterations (N = 594). P = 0.0002; log-rank (Mantel–Cox) test.