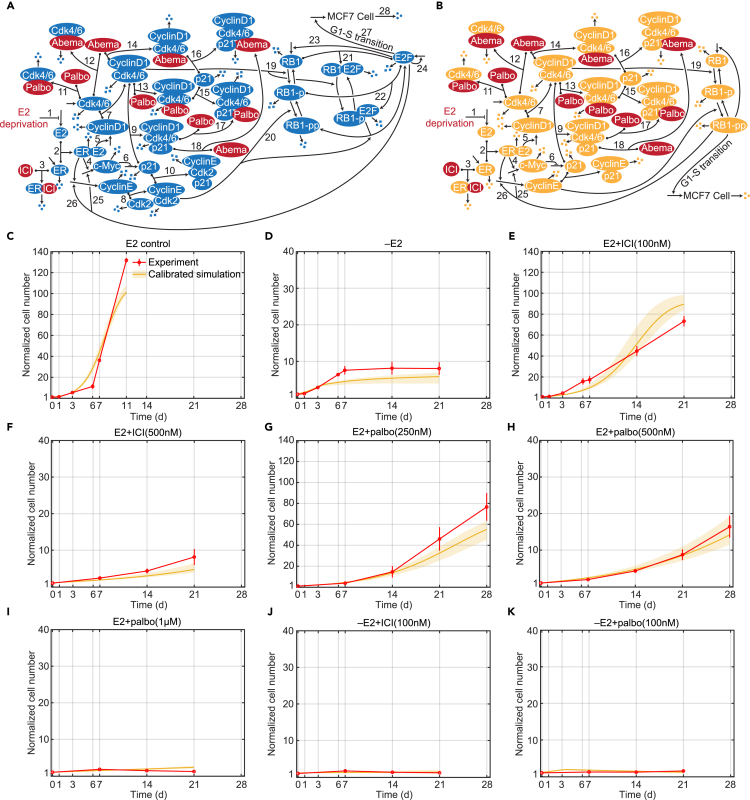
Figure 1.
Signaling diagram of the biological mechanism, model structure and model calibrations
(A) Detailed reactions of the biological mechanism related to estrogen signaling and Cdk4/6 inhibition. Reversible binding reactions are represented by dots on the components and an arrow to the complex. Three dots represent degradation of a protein or the death of a cell. Arrows pointing from blank space to a protein or MCF7 cell represent production of the protein or proliferation of the cell. Arrows pointing from one protein to another protein represent phosphorylation or dephosphorylation of the protein. Lines pointing to other lines represent enhancement (arrow) or inhibition (blunt head) of the reactions. Treatments are colored red. The numbered biological mechanism consisting of the following process: 1. –E2 decreases estrogen; 2. E2 binds to ER; 3. ICI binds to ER; 3. E2:ER increases transcription of c-Myc; 5. E2:ER increases transcription of cyclinD1; 6. c-Myc inhibits transcription of p21; 7. CyclinD1 binds to Cdk4/6; 8. CyclinE binds to Cdk2; 9. p21 binds to cyclinD1:Cdk4/6; 10. p21 binds to cyclinE:Cdk2; 11. Palbociclib binds to Cdk4/6; 12. Abemaciclib binds to Cdk4/6; 13. Palbociclib binds to cyclinD1:Cdk4/6; 14. Abemaciclib binds to cyclinD1:Cdk4/6; 15. p21 binds to cyclinD1:Cdk4/6:palbociclib; 16. p21 binds to cyclinD1:Cdk4/6:abemaciclib; 17. Palbociclib binds to cyclinD1:Cdk4/6:p21; 18. Abemaciclib binds to cyclinD1:Cdk4/6:p21; 19. CyclinD1:Cdk4/6 phosphorylates RB1; 20. CyclinE:Cdk2 phosphorylates RB1-p; 21. RB1 binds to E2F; 22. RB1-p binds to E2F; 23. E2F up-regulates RB1; 24. E2F up-regulates itself; 25. E2F up-regulates c-Myc; 26. E2F up-regulates cyclinE; 27. E2F drives the G1-S cell cycle transition and proliferation; 28. Cell death. (STAR Methods).
(B) Structure of the mathematical model, a simplified version of the biological mechanism in (A).
(C) Model calibration to experimental data (mean ± s.e., n = 3) in E2 control condition. The experimental data are shown in red and the calibration simulation results are shown in yellow (solid line represents the lowest cost value simulation and the shaded regions contains the central 98% of the cohort simulations).
(D) Model calibration to experimental data (mean ± s.e., n = 3) in –E2 condition.
(E) Model calibration to experimental data (mean ± s.e., n = 3) in E2+ICI(100 nM) condition.
(F) Model calibration to experimental data (mean ± s.e., n = 3) in E2+ICI(500 nM) condition.
(G) Model calibration to experimental data (mean ± s.e., n = 3) in E2+palbo(250 nM) condition.
(H) Model calibration to experimental data (mean ± s.e., n = 3) in E2+palbo(500 nM) condition.
(I) Model calibration to experimental data (mean ± s.e., n = 3) in E2+palbo(1 μM) condition.
(J) Model calibration to experimental data (mean ± s.e., n = 3) in –E2+ICI(100 nM) condition.
(K) Model calibration to experimental data (mean ± s.e., n = 3) in E2+palbo(100 nM) condition.