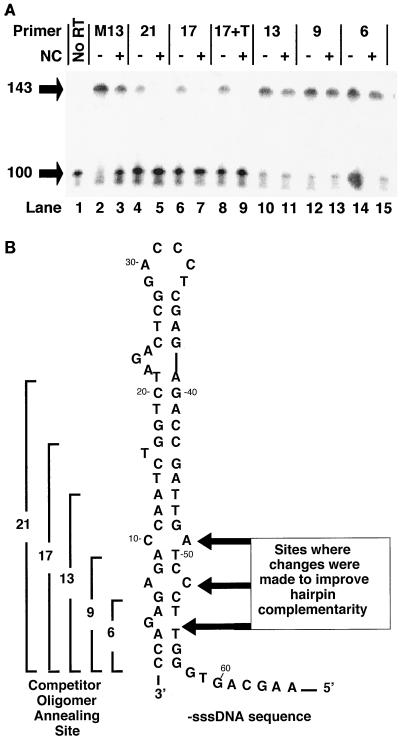
FIG. 4.
NC blocks self-priming by promoting the annealing of complementary oligomers that are more stable than the hairpin. (A) A synthetic oligonucleotide comprising the 3′ 100 bases of the −sssDNA was end labeled, heated, and slowly cooled in the presence of either a 21-nt M13 DNA primer (lanes 2 and 3) or DNA competitor of decreasing length, complementary to the 3′ end of the −sssDNA (lanes 4 to 15; see panel B). The length of each oligonucleotide is indicated at the top, and the site of annealing to the hairpin is shown in panel B. 17+T has the same sequence as 17, with the addition of a 7-nt unannealed tail at the 3′ end (see text). Each reaction was performed both in the presence and in the absence of enough NC to coat the primer-template at 7 bases/NC, as indicated at the top. Positions of migration of the 100-nt −sssDNA and the 143-nt self-primed product are shown at left. Lane 1 contains unmodified, labeled 100-base DNA. (B) The −sssDNA sequence which folds into a structure resembling the TAR hairpin. The lengths and sites of annealing of the competitor DNA oligomers discussed above are shown at the left. The sequence of the competitor DNA oligomer used to block self-priming is identical to the genomic RNA sequence. The sites of modifications of the hairpin discussed in the text are indicated by arrows at the right.