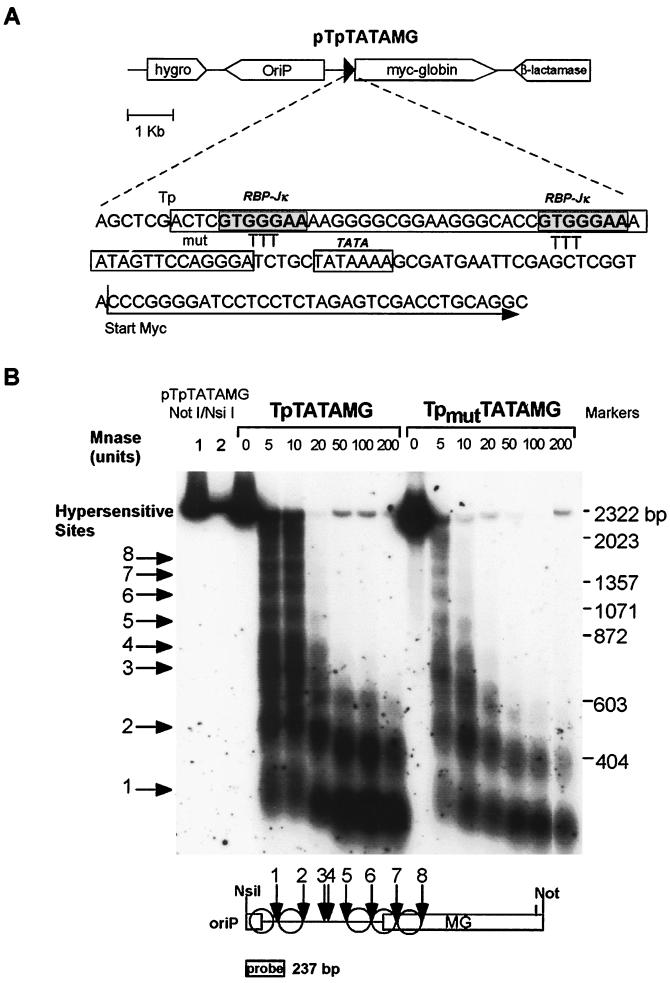
FIG. 1.
Schematic representation of pTpTATAMG and micrococcal nuclease digestion of pTpTATAMG and pTpmutTATAMG. (A) Main features of plasmid pTpTATAMG. The sequence of the EBV TP1/LMP2A promoter, TATA box, upstream Myc-globin reporter gene, and heptameric RBP-Jκ/CBF1 binding sites within the TP1/LMP2A promoter are indicated. Substitutions in plasmid pTpmutTATAMG (mut) replace the GGG sequence within the RPB-Jκ site with TTT. (B) Micrococcal nuclease (Mnase) digestion and indirect end labeling of the upstream region of the Myc-globin reporter gene. The probe for hybridization was derived from the 237-bp NsiI/BamHI fragment of pTpTATAMG (position indicated). The eight micrococcal nuclease-hypersensitive sites identified are numbered 1 to 8. Proposed positions of the nucleosomes (circles) are depicted in the lower panel. Marker lanes (1 and 2) contain 10 and 2 pg of pTpTATAMG digested with NotI and NsiI.