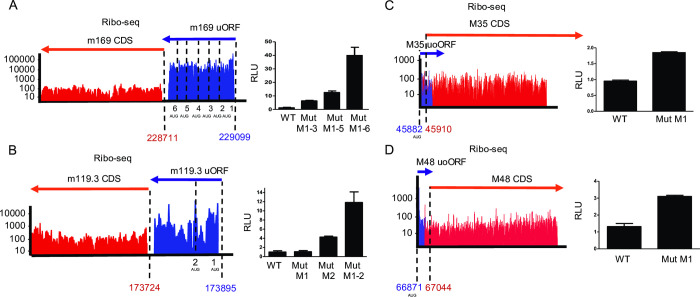
Fig 9. MCMV uORFs/uoORFs tune viral gene expression.
Ribo-seq data (aggregated reads in logarithmic scale) of the respective viral genomic loci and their validation by dual luciferase assays are shown for m169 uORF (A), m119.3 uORF (B), M35 uoORF (C) and M48 uoORF (D). The number of AUG codons for the respective viral u(o)ORFs are indicated. Coordinates represent the start codons of the u(o)ORFs and ORFs. Both ORFs and u(o)ORFs were annotated based on translational start site profiling data, including Harringtonine and Lactimidomycin pre-treated samples and PRICE analysis. Manual curation assessed the presence of upstream TiSS, start codon usage and presence of STOP codons to identify relevant ORFs. psiCheck-2 reporter plasmids harbored the indicated MCMV u(o)ORFs (WT) and AUG start codon mutants thereof (Mut) upstream of firefly-luc reporter gene. Luciferase assay data at 48h post transfection are shown as mean RLU (Firefly/Renilla ratio) of three biological replicates (n = 3) plotted along with the standard error (S.E.M.).