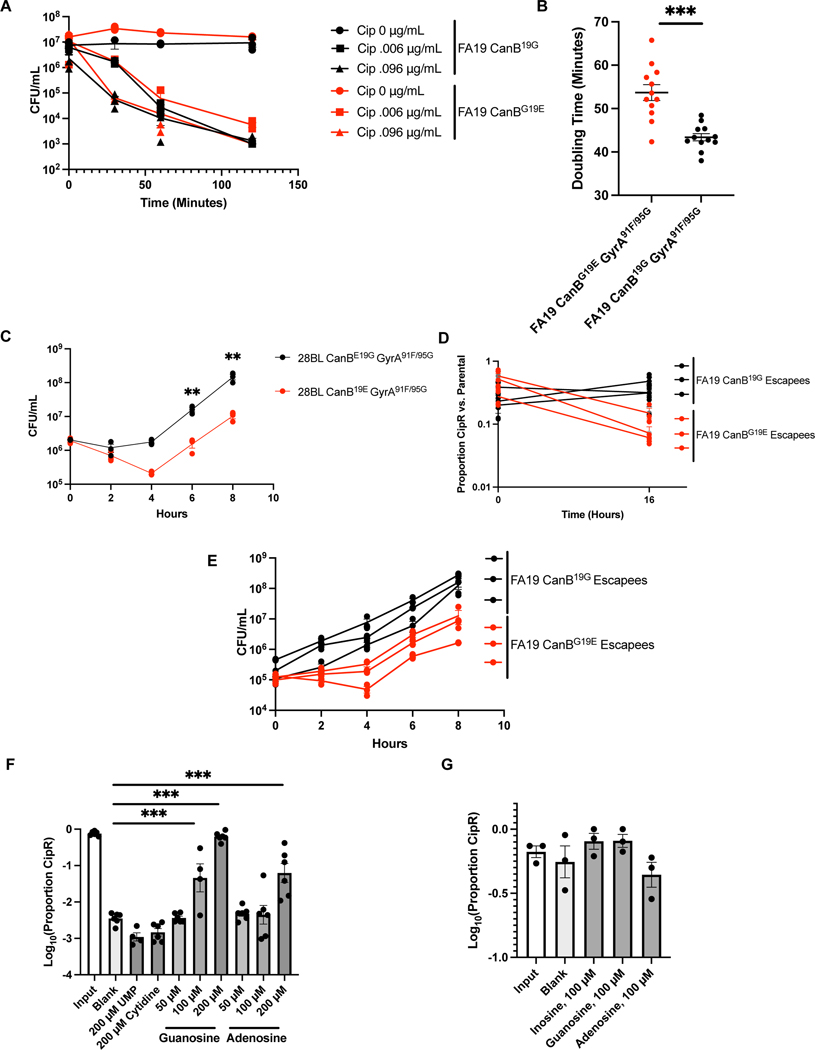
Extended Data Fig. 7.
The CanB19G variant does not affect killing by ciprofloxacin, but CanB19G provides an advantage in the presence of gyrA mutations across multiple gyrA alleles and strain backgrounds. (A) Kill curve of isogenic CanB FA19 at 2x MIC and 32x MIC (N=3, representative of two independent experiments). (B) Doubling time of replicates in Fig. 3E as determined by linear regression on log-transformed CFU/mL counts from 2–10 hours of growth (N=12, from two independent experiments, error bars represent SEM). Significance (p = 1.9e-5) by unpaired two-sided t-test. (C) Growth of CanB isogenic isogenic 28BL strains bearing ciprofloxacin resistance-determining GyrA91F/95G (N=3, representative of two independent experiments). Significance determined by unpaired two-sided t-test. (D) Competition between spontaneously ciprofloxacin resistant isogenic CanB strains (see Extended Data 2) and susceptible parental strains (N=3, representative of two independent experiments). (E) Growth curves of CanB isogenic FA19 strains with spontaneous ciprofloxacin resistance-determining gyrA alleles (N=3, representative of two independent experiments). (F and G) Competition between (F) FA19 CanBG19E and FA19 CanBG19E GyrA91F (N=6, except UMP and 100 μM Adenosine, N=4) and (G) FA19 CanB19G and FA19 CanB19G GyrA91F (N=3) after 16 hours along with media supplementation in liquid GCP-K (representative of two independent experiments, error bars represent SEM). Significance (from left to right, p = 0.0002, p = 5.7e-13, p = 3.86e-5), by oneway ANOVA with Dunnett’s multiple comparisons test. *p<0.05, **p<0.01, ***p<0.001