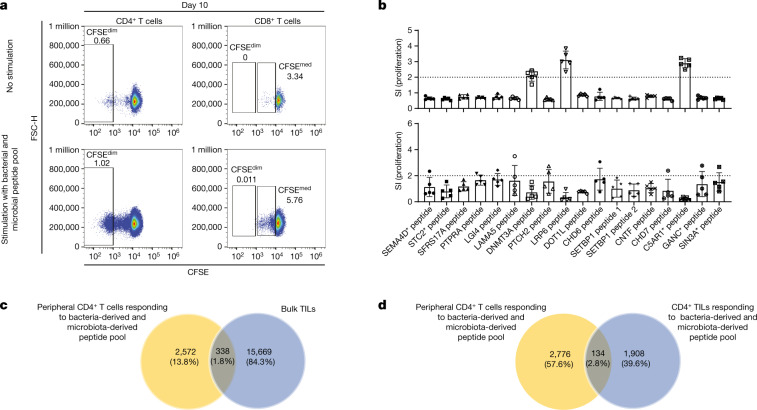
Fig. 5. Peripheral memory T cells recognize several tumour antigens after activation with bacteria-derived and microbiota-derived peptides.
a, CD45RA− peripheral T cells were isolated and labelled with CFSE. Cells were then stimulated with the bacteria-derived and microbiota-derived peptide pool for 10 days. Next, 60 replicate wells were pooled and proliferative (CFSEdim) CD4+ T cells were sorted and short-term expanded. b, Bacteria-reactive or microbiota-reactive peripheral CD4+ T cells responded to several tumour antigens from the autologous tumour when stimulated with peptide presented by irradiated autologous PBMCs as APCs (top). Proliferation was measured after 5 days by 3H-thymidine incorporation (data represent the means of five replicate wells ± s.e.m.). Peripheral blood memory T cells were isolated and directly tested with tumour antigens (bottom). Proliferation was measured after 5 days by 3H-thymidine incorporation (data represent the means of five replicate wells ± s.e.m.). c,d, Part of CFSEdim (that is, proliferating) peripheral CD4+ T cells that responded to the bacteria-derived and microbiota-derived peptide pool was used for TCRVβ sequencing. The percentage and frequency of TCR overlap of unique productive TCRVβ sequences of bacteria-reactive and microbiota-reactive peripheral CD4+ (CFSEdim) and TCRVβ sequences of bulk TILs (c), and the overlap of unique productive TCRVβ sequences of bacteria-reactive and microbiota-reactive peripheral CD4+ (CFSEdim) and TCRVβ sequences of (CFSEdim) bacteria-reactive and microbiota-reactive TILs (d) are shown. Also see Extended Data Figs. 6 and 7.