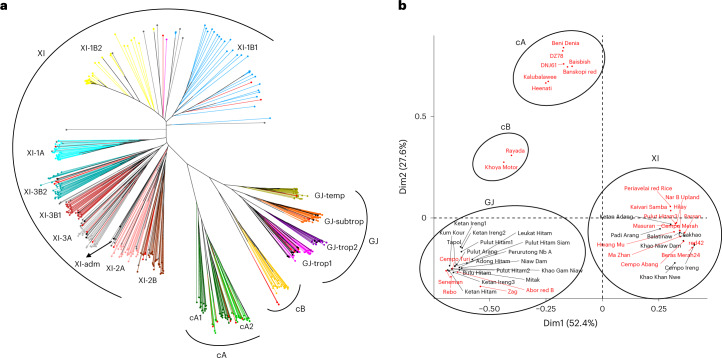
Fig. 1. Population genomic analysis of pigmented rice.
a, Neighbour-joining tree for K = 15 subpopulations and one admixture group. The phenogram shows the clustering of the 474 accessions selected from the 3K-RG dataset (clockwise: GJ-temperate (okra), GJ-subtropical (orange), GJ-tropical2 (dark magenta), GJ-tropical1 (magenta), cB (goldenrod), cA1 (dark green), cA2 (light green), XI-2B (brown), XI-2A (pink), XI-adm (dark grey), XI-3A (light grey), XI-3B1 (brick red), XI-3B2 (turquoise), XI-1A (cyan), XI-1B2 (yellow) and XI-1B1 (sky blue)) and the 51 pigmented varieties (the red and black branches represent red- or black-pigmented varieties, respectively). O. sativa subpopulations are defined as described by Zhou et al.14. b, Principal component analysis plot showing the clustering of the pigmented varieties into the four main groups of O. sativa. The analysis was performed on 51 pigmented varieties and 474 accessions selected from the 3K-RG dataset, but only the 51 pigmented varieties are shown. Red and black names represent red and black pigmented varieties, respectively.