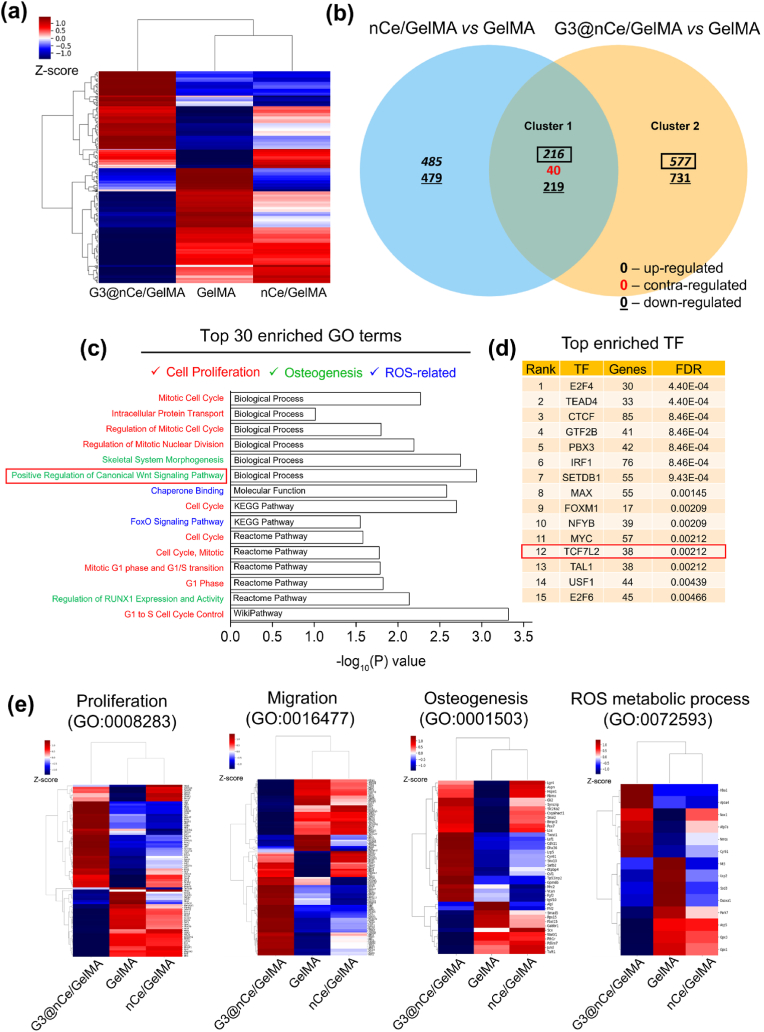
Fig. 6.
Transcriptome analysis reveals the key functions and signalling pathways activated by G3@nCe/GelMA. (a) The distance-based clustering analysis from global transcriptional changes in GelMA, nCe/GelMA and G3@nCe/GelMA groups with 1.5 fold change and over 2 log2 values from the combinative comparison. (b) Venn diagram showing the comparison between differential gene expression across 2 key comparisons (nCe/GelMA vs GelMA and G3@nCe/GelMA vs GelMA) based on co-up or –down and contra-regulated genes. (c) Among the top 30 enriched GO or pathway using up-regulated genes from cluster 1 (mainly due to the presence of nanoceria) using DAVID (BP, MF, KEGG, Reactome and Wikipathway), major terms related to cell proliferation, osteogenesis and ROS-related processes were disclosed. (d) Transcription factor enrichment (TFE) of cluster 1 was analyzed using ChEA3, a web-based tool indicating the enrichment of TCF7L2, a key transcription factor involved in Wnt signalling pathway. (e) Heat map showing DEGs in G3@nCe/GelMA, nCe/GelMA, and pristine GelMA from specific GO terms related to cell proliferation, migration, osteogenesis, and ROS-metabolic process.