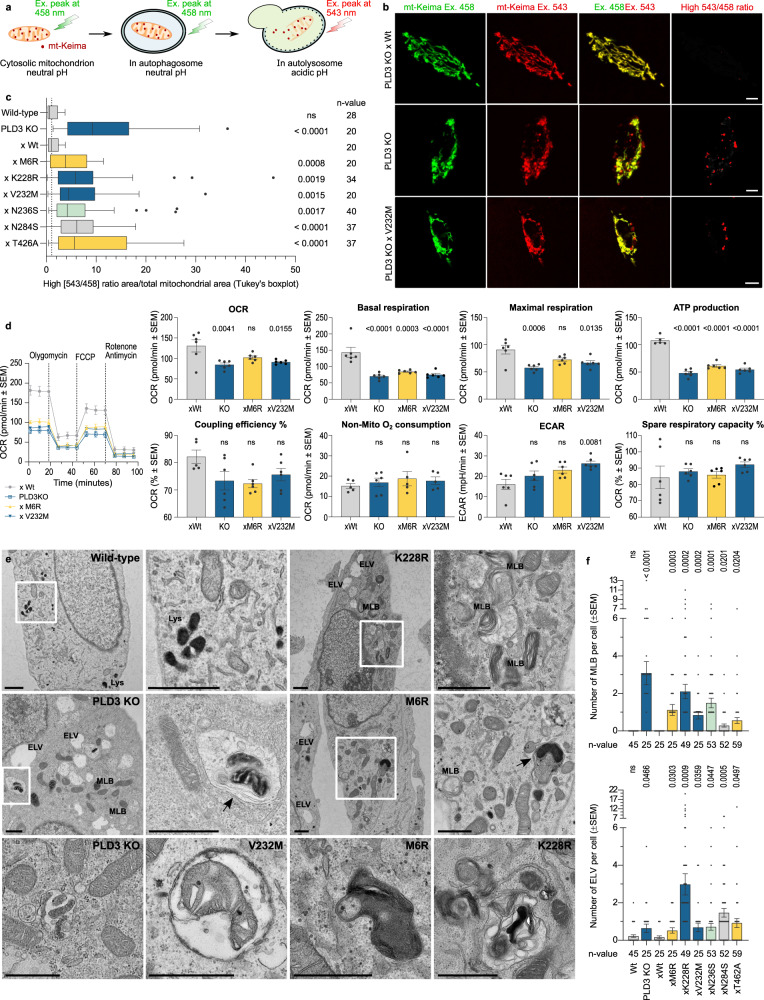
Fig. 2. Mitochondrial fitness and clearance are disrupted by PLD3 dysfunction.
a Schematic representation of Mt-Keima, showing a pH-dependent shift in fluorescence excitation that can be imaged in two channels. b Representative confocal images of SH-SY5Y PLD3 cell models. Scale bar = 5 µm. c Tukey’s boxplot (first to third quartile box with the median as a horizontal line) showing the index of mitophagy, which was calculated as the parameter: high [543/458] ratio area/total mitochondrial area (n = 20–40). Two-tailed, unpaired t-tests were used for statistical testing. Differences with the wild-type rescue condition are indicated. d Mitochondrial bioenergetic profiles were obtained with the Seahorse XFp Cell Mito Stress test, including oxygen consumption rate (OCR) and extracellular acidification rate (ECAR). An ordinary one-way ANOVA with Bonferroni’s multiple comparisons test was used for statistical testing (n = 6; columns show mean values ± SEM). e Representative EM images of PLD3 KO and SNP variant-rescued SH-SY5Y cells showing an aberrant accumulation of degradative organelles, including MLBs and autophagosomes/autophagolysosomes, often with remnants of mitochondria. ELV electron-lucent vesicles, MLB multilamellar bodies. Scale bar is 1 µm. f Quantification of the number of MLB and ELV per cell in two PLD3 clonal knockout lines, of which results were pooled together. Statistical significance was tested in comparison to the wild-type rescue values using a two-tailed, unpaired t-test (n = 25–59). Source data are provided as a Source Data file.