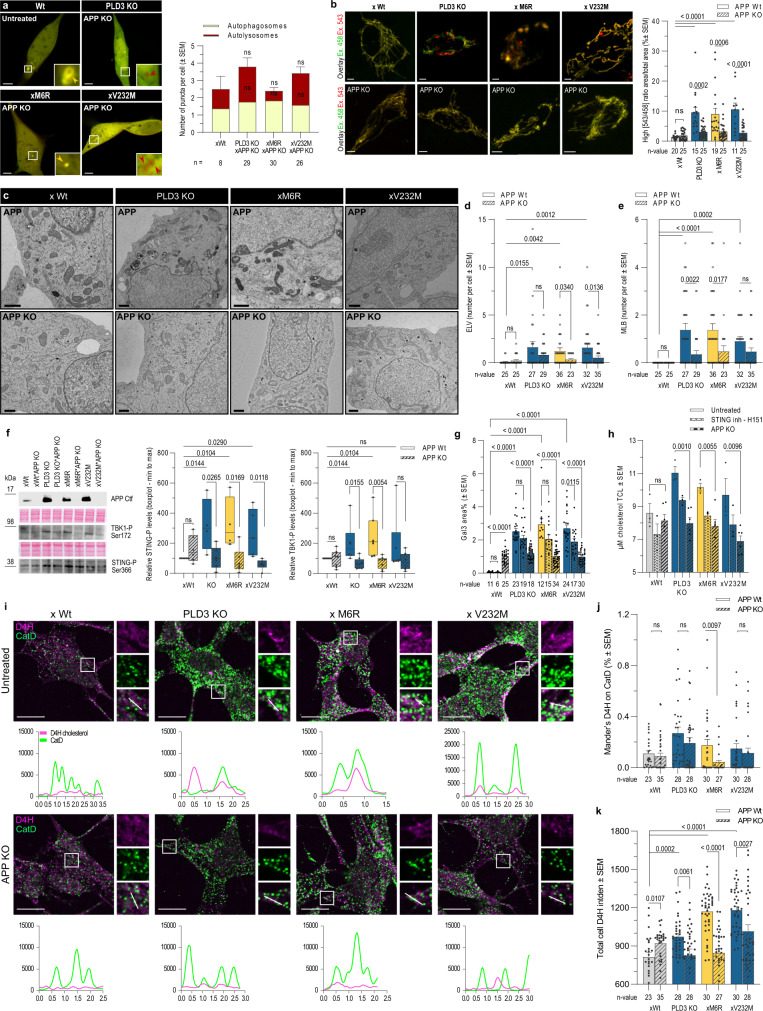
Fig. 7. APP depletion and STING inhibition ameliorate endolysosomal pathology upon PLD3 dysfunction.
a Representative airyscan images of autophagosome/-lysosome puncta as in Fig. 6b, using SH-SY5Y knocked out for APP. These show LC3 puncta levels indistinguishable from xWt levels. Scale bar = 5 µm. Statistical significance was calculated using two-tailed, unpaired t-tests (specific n-values are listed on the graph). b The index of mitophagy in APP KO cells was calculated using mt-Keima (scale bar = 5 µm). The graph shows a high [543/458] ratio area/total mitochondrial area. An ordinary one-way ANOVA with Bonferroni’s multiple comparisons test was used (n = 11–25). c Representative transmission electron microscopy (TEM) images showing a rescue effect of an APP KO in a PLD3 dysfunction background. Counts of d electron-lucent vesicles (ELV) and e multilamellar bodies (MLB) per cell as identified on TEM images (n = 25–36). Scale bar = 1 µm. Two-tailed, unpaired t-tests were used for statistical testing. f APP knockout double transgenics were analyzed on western blot for STING activation (n = 6) and TBK1 autophosphorylation at Ser172 (n = 8). Statistical differences were calculated using two-tailed, unpaired t-tests and depicted on boxplots (min to max; median as the center, the box extends from the 25th to 75th percentiles, and whiskers range from the smallest to the greatest value). g Quantification of Gal3 levels on confocal images (n = 6–34). Statistical significance was calculated using an ordinary one-way ANOVA with Bonferroni’s multiple comparisons test. h Beneficial effect on cholesterol levels in total cell lysates (TCL; n = 3 in untreated and H151 conditions, n = 6 in the APP KO conditions) after 5 h H151 treatment or after an APP KO, using the Cholesterol Glo Assay. Statistical differences were calculated using two-tailed, unpaired t-tests. i Top: representative confocal images of untreated and APP knockout SH-SY5Y cells, stained with the D4H-cholesterol probe (magenta) and CatD (green). Images were taken in confocal mode on a ZEISS LSM 900. Scale bar = 10 µm. Bottom: Co-localization analysis using line intensity profiles. Quantification of IF images using j Manders’ coefficient and k integrated densities per cellular area (n = 23–35). Statistical differences were calculated using two-tailed, unpaired t-tests as indicated. Source data are provided as a Source Data file.