Abstract
Circulating levels of the astrocytic marker S100B have been associated with risk of neuropsychiatric or neurological disorders. However, reported effects have been inconsistent, and no causal relations have yet been established. We applied two-sample Mendelian Randomization (MR) on the association statistics from genome-wide association studies (GWAS) for circulating S100B levels measured 5-7 days after birth (the iPSYCH sample) and in an older adult sample (mean age, 72.5 years; the Lothian sample), upon those derived from major depression disorder (MDD), schizophrenia (SCZ), bipolar disorder (BIP), autism spectral disorder (ASD), Alzheimer’s disease (AD), and Parkinson’s disease (PD). We studied the causal relations in the two S100B datasets for risk of these six neuropsychiatric disorders. MR suggested increased S100B levels 5-7 days after birth to causally increase the risk of MDD (OR = 1.014; 95%CI = 1.007–1.022; FDR-corrected p = 6.43×10−4). In older adults, MR suggested increased S100B levels to have a causal relation to the risk of BIP (OR = 1.075; 95%CI = 1.026–1.127; FDR-corrected p = 1.35×10−2). No significant causal relations were found for the other five disorders. We did not observe any evidence for reverse causality of these neuropsychiatric or neurological disorders on altered S100B levels. Sensitivity analyses using more stringent SNP-selection criteria and three alternative MR models suggested the results are robust. Altogether, our findings imply a small cause-effect relation for the previously reported associations of S100B and mood disorders. Such findings may provide a novel avenue for the diagnosis and management of disorders.
Subject terms: Depression, Predictive markers
Background
Neuropsychiatric and neurological disorders are debilitating diseases with relatively high lifetime prevalence, incurring large burdens on families and society [1]. However, the causes of neuropsychiatric disorders are largely unknown. Recent large-scale GWAS have revealed a large number of genomic regions that associate with one or more of the mental disorders [2–7], but the exact causal genes underlying these disorders have proved hard to identify due to complex linkage disequilibrium (LD) in the genome. Nevertheless, aggregating association signals into biological pathways has uncovered several pathways that play key roles in the pathophysiology of mental disorders, such as synaptic regulations, neuroinflammation and glucose metabolism [2–7]. These pathways implicate not only neuronal but also glial cell dysfunction as a cause of neuropsychiatric disorders [8, 9].
The calcium-binding protein—S100B—is a classic marker for activated astrocytes. S100B levels in peripheral blood have been suggested as a biomarker for neuropsychiatric and neurological disorders [10, 11]. Although knowledge on the definitive biophysiological functions of S100B is incomplete, it has been demonstrated that S100B could contribute to intracellular and extracellular calcium homeostasis, cell survival and proliferation, and other enzymatic bioprocesses [12]. On the one hand, elevated circulating levels of S100B have been associated with risk of Schizophrenia (SCZ), bipolar disorders (BIP), major depression disorder (MDD), autism spectral disorders (ASD), and Alzheimer’s disease (AD) [11, 13–18]. However, others have also reported findings in the opposite direction [19, 20], or null associations [21]. As S100B levels have been shown to increase with age and be altered by the use of certain medications [22–24], differences in studied subjects may partly underlie conflicting results. Currently, it is not known whether alterations in blood S100B levels are causes or consequences of these disorders.
Mendelian randomization (MR) is a recently well-developed statistical framework for interrogating causal relations between an exposure and outcome [25, 26], based on observational data. Within this framework, the exposure variables of interest are indirectly randomized by ‘nature’ through genetic instrument variables (mostly single nucleotide polymorphisms (SNPs)) that strongly associate with the variation of exposures. Under certain assumptions, the average causal effects of exposures on outcomes can be estimated [27]. This framework is especially powerful if combined with the association statistics from large-scale GWAS using two-sample MR models [28]. The advantages of this include accurate effect size estimation using extremely large samples, and reduced confounder bias through measuring both exposure and outcomes in the same subjects.
Here, we present the first study investigating the causal effect of circulating S100B levels on the risk of six major neuropsychiatric or neurological disorders. We applied two-sample MR methods on the recently published large-scale GWAS for blood S100B levels measured 5-7 days after birth (N = 8138; the iPSYCH dataset) [29] and in an older adult sample (mean age: 72.5 years; N = 769; the Lothian dataset) [30, 31] as exposure, and case-control GWAS for SCZ (N = 79,845) [2], BIP(N = 51,710) [3], MDD(N = 500,199) [32], ASD(N = 46,350) [7], AD(N = 63,976), and Parkinson’s Disease (PD) (N = 482,730) [33] as outcomes. We further confirmed our findings by performing a range of sensitivity analysis varying the number of genetic instruments and using alternative MR models.
Methods
Study Aim and Design
The aim of the present study was to examine whether circulating levels of S100B are causally associated with the risk of neuropsychiatric disorders. To achieve this aim, we used an open-source two-sample MR program [28]. We took the association statistics of SNPs for our exposure – here S100B – and those for our outcomes SCZ, BIP, MDD, ASD, AD, and PD, as input. SNP association statistics were obtained from publicly accessible databases (see Availability of data and materials). This study follows the STROBE-MR guidelines [34].
Exposure GWAS
GWAS for the circulating S100B levels were performed on dried blood spots samples collected 5-7 days after birth by the iPSYCH project [29, 35]. Levels of S100B were determined using the Meso-scale platform [36]. All participants to the GWAS are Danish, and 8,138 subjects in total were included in the GWAS. During the association analysis, age at 2012, sex and six genetic principal components (PCs) were included as covariates. The effects of SNPs were reported on the standard deviation of the log-transformed S100B levels in dried blood spots (1 SD = 3043 pg/mL). The second set of exposure statistics were derived from a GWAS of the Lothian Birth Cohort 1936, which included 769 participants (mean age 72.5 years). S100B levels from the blood for this sample were measured by the chemiluminescence immunoassay S100B kit (catalogue number 314701, distributed by DiaSorin, Berks, UK) [30]. The measured S100B levels were rank-based inverse transformed, and the effects of age, sex, and body mass index were regressed out before running GWAS [30].
Outcome GWAS
GWAS results for the four psychiatric disorders were obtained with permission from the Psychiatric Genomics Consortium (PGC), each of which followed a case-control design and is an inverse-variance weighted meta-analysis across tens of participating GWAS sub-studies. All GWAS of sub-studies were performed using the Ricopili pipeline [37], thereby maintaining a consistent analysis protocol. For each sub-study GWAS, sex, age, and genetic principal components, the number of which varied between sub-studies, were included as covariates. Since our exposure GWAS were performed on mainly European populations, we only used GWAS results derived primarily from European ancestral samples, to reduce bias induced by ancestral differences. In addition, for the MDD GWAS, we used the dataset excluding samples from 23andMe.
The AD GWAS results were obtained through online application to the International Genomics of Alzheimer’s Project (IGAP) [5]. IGAP is a large three-stage study based upon genome-wide association studies (GWAS) on individuals of European ancestry. In stage 1, IGAP used genotyped and imputed data on 11,480,632 single nucleotide polymorphisms (SNPs) to meta-analyse GWAS datasets consisting of 21,982 Alzheimer’s disease cases and 41,944 cognitively normal controls from four consortia: The Alzheimer Disease Genetics Consortium (ADGC); The European Alzheimer’s disease Initiative (EADI); The Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium (CHARGE); and The Genetic and Environmental Risk in AD Consortium Genetic and Environmental Risk in AD/Defining Genetic, Polygenic and Environmental Risk for Alzheimer’s Disease Consortium (GERAD/PERADES). In stage 2,11,632 SNPs were genotyped and tested for association in an independent set of 8362 Alzheimer’s disease cases and 10,483 controls. Meta-analysis of variants selected for analysis in stage 3 A (n = 11,666) or stage 3B (n = 30,511) samples brought the final sample to 35,274 clinical and autopsy-documented Alzheimer’s disease cases and 59,163 controls. In the present study, the stage 1 data was used.
The PD GWAS results were obtained from the fixed-effect meta-analysis performed by the International Parkinson Disease Genomics Consortium (IPDGC) [33]. This study included both patients and proxy-patients (i.e. those who were not diagnosed but their first-degree relatives were) as cases. A similar GWAS protocol was used by each sub-study before the meta-analysis. All individual sub-studies had adjusted for sex, age, and up to 10 PCs in their GWAS (see Nalls et al. [33]). In the present study, we used the association results excluding 23andMe samples.
Instrumental SNPs selection
Prior to selecting instrumental SNPs, we quality checked the downloaded GWAS summary statistics. We removed variants with minor allele frequencies (MAF) < 0.05, and those that are not biallelic in the 1000 Genomes Project Phase 3 European samples(1KGP3), from all GWAS datasets. In addition, we removed SNPs that are ambiguous in allelic coding, i.e., A/T or C/G or were imputed with INFO score <0.6 when this information was available. To ensure comparability across disorders in terms of MR results, we removed SNPs that only exist in subsets of the GWAS results.
To select uncorrelated instrumental SNPs, we used PLINK [38] and the LD structure of the 1KGP3 dataset. The following parameters in PLINK were set, clump-kb 10,000 kb, -clump-p1 10−6, and -clump-r2 0.05. The association statistics for the instrumental SNPs to S100B and to each of the disorders were harmonized by assigning the same allele as the effective allele. The instrumental strengths of SNPs to S100B level was evaluated using the F-statistic [39]. SNPs having F-statistics >10 were considered as candidate for instruments. A full list of SNPs selected as instruments are presented in Supplementary Tables 1-30.
Two-sample MR analysis
To test whether S100B levels have a causal effect on neuropsychiatric or neurological outcomes, we performed a two-sample MR analysis using S100B levels as exposure and each neuropsychiatric or neurological disorder as the outcome. To mitigate bias induced by horizontal pleiotropy (i.e., same genetic variants affecting both the exposure and outcome but through independent pathways) we first detected horizontal pleiotropic SNPs using the Mendelian Randomization Pleiotropy RESidual Sum and Outlier (MR-PRESSO) method [40]. Specifically, we used the MR-PRESSO global test to check whether there was horizontal pleiotropy in the instrumental SNP set; if the test was significant (p < 0.05), we used the MR-PRESSO outlier test to identify the pleiotropic SNPs and remove them from subsequent analysis. For our primary analysis we used the powerful invariance-weighted MR model (IVW) to detect causal effects. The Benjamini-Hochberg procedure was used to adjust multiple testing across disorders [41].
We then tested the possibility of reverse causality, namely that neuropsychiatric or neurological disorders have a causal effect on S100B levels, using each of the six disorders as exposure and S100B as the outcome in IVW-MR analysis.
Sensitivity analysis
To confirm our results, we additionally applied three other two-sample MR methods: the MR-Egger regression, weighted median, and the MR robust adjusted profile score (MR-RAPS) [26]. To test if our results were due to the use of subgenome-wide significant SNPs (p < 10−6), we reran our analysis using SNPs that met the standard significance level, i.e., p < 5×10−8.
Results
We detected horizontal pleiotropy between S100B levels and SCZ (p < 0.001), BIP (p < 0.01) and AD (p = 0.02), based on the MR-PRESSO global test. To mitigate bias induced by this, we removed SNPs flagged as pleiotropic (uncorrected p < 0.05) for SCZ (6 SNPs), BIP (2 SNPs), and AD (2 SNPS) using the MR-PRESSO outlier test. After removing these, we did not observe any evidence of horizontal pleiotropy (p > 0.05).
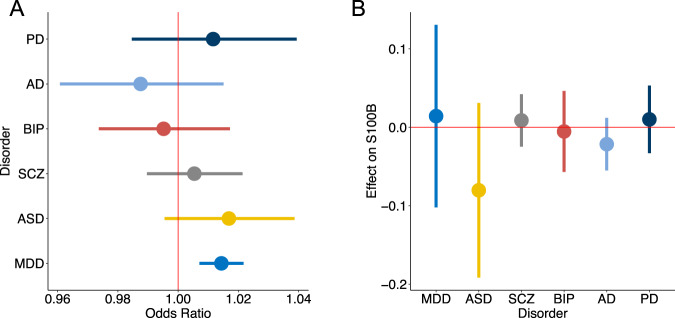
We then applied IVW-MR on each of the six neuropsychiatric or neurological disorders. Figure 1 shows that increased S100B levels 5–7 days after birth were causally associated with increased risk of MDD (Odds ratio (OR) = 1.014; 95%CI = 1.007–1.022; FDR-corrected p = 6.43×10−4). We detected no significant causal effects of S100B on the risk to SCZ, BIP, ASD, AD and PD (p > 0.05, Table 1). The significant causal effect of S100B on MDD was also detected by the MR-RAPs and weighted median methods, both of which showed very similar effect sizes (Supplementary Table 31), whereas the Egger-regression method showed only a trend (OR = 1.012, 95%CI = 0.9988–1.027; uncorrected p = 0.0835). Using more significant instrumental SNPs (association p < 5×10−8), we again found significant causal effects of S100B on MDD by IVW, MR-RAPs and weighted median methods (Supplementary Table 32). For the other five neuropsychiatric or neurological disorders, we again detected no significant causal relations for any of these three methods, also at the stringent association threshold.
Fig. 1. Bidirectional causal relations between S100B (iPSYCH) and six neuropsychiatric disorders.
A Inverse variance weighted (IVW) MR models were used to estimate average causal effect of circulating levels of S100B measured in the iPSYCH sample on six neuropsychiatric disorders. Odds ratios and 95% confidence intervals are shown. B Causal effect of six neuropsychiatric disorders on S100B levels. Average effect size (beta) and 95% confidence intervals are shown. PD Parkinson’s disease, AD Alzheimer’s disease, BIP bipolar disorder, SCZ Schizophrenia, ASD autism spectral disorders, MDD major depression disorder.
Table 1.
Causal effects of S100B levels measure at birth on neuropsychiatric disorders.
| Outcome | NSNP | OR | OR_95%CI | Het_p | MR-PRESSO | pval | padj |
|---|---|---|---|---|---|---|---|
| SCZ | 34 | 1.005 | 0.990–1.021 | 0.580 | 0.613 | 0.504 | 1.0 |
| BIP | 32 | 0.995 | 0.974–1.017 | 0.454 | 0.347 | 0.666 | 1.0 |
| ASD | 37 | 1.017 | 0.996–1.039 | 0.294 | 0.362 | 0.122 | 0.612 |
| MDD | 33 | 1.014 | 1.007–1.022 | 0.162 | 0.248 | 1.07 × 10−4 | 6.43 × 10−4 |
| AD | 44 | 0.988 | 0.961–1.015 | 0.109 | 0.121 | 0.372 | 1.0 |
| PD | 40 | 1.012 | 0.985–1.039 | 0.433 | 0.448 | 0.402 | 1.0 |
SCZ Schizophrenia, BIP bipolar disorder, ASD autism spectral disorders, MDD major depression disorder, AD Alzheimer’s disease, PD Parkinson’s disease, NSNP number of instrumental SNPs selected, OR odds ratio for disorders, OR_95%CI the 95% confidence interval for OR, Het_p p value of the test of heterogeneity of instrumental SNP, MR_PRESSO p value of the global horizontal pleiotropy test, pval p value for the MR analysis, padj Benjamini-Hochberg False discovery rate adjusted p value.
The causal effects of S100B level measure in the iPSYCH sample on six neuropsychiatric disorders were estimated by the Inverse Variance Weighted MR method. Instrumental SNPs for S100B were selected from SNPs that associates with S100B levels at p < 10−6.
In the reversed direction, we did not find any causal effects of neuropsychiatric or neurological disorders on S100B levels 5-7 days after birth (Fig. 1B). Neither did the other three MR methods find any relations (Supplementary Table 33). Using the stringent association threshold for instrumental SNPs (p < 5 × 10−8) still gave the same negative results (Supplementary Table 34).
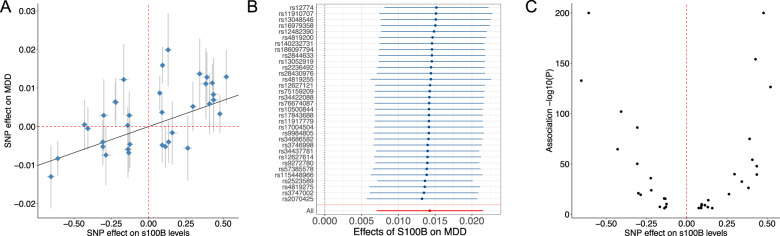
We then investigated the details of the causal effect of S100B on MDD risk (see Fig. 2). We found that a one-standard deviation (SD) increase in S100B levels measured 5-7 days after birth increases the risk of MDD later in life by an odds ratio of 1.014 (95%CI = 1.007–1.022; Fig. 2A). The instrumental SNPs used to detect this relation were all consistent in direction of effect (as shown in the leave-one-out analysis (Fig. 2B)), did not show significant heterogeneity in the effect of S100B on MDD (Cohran’s heterogeneity, p = 0.162), and their effect on S100B levels were close to symmetric around the zero line (Fig. 2C). These diagnostic results imply the causal effect of S100B on MDD is reliable.
Fig. 2. Effects of instrumental SNPs on MDD and S100B (iPSYCH).
A Effects of selected instrumental SNPs on MDD (on the scale of log (odds ratio)) and on S100B levels (on the scale of standard deviation of S100B levels) measured in the iPSYCH sample. Confidence interval (95%) are also shown. B IVW estimates for the effect of S100B on MDD after excluding each of the SNPs on the y axis, i.e., leave-one-out. The overall effect is indicated at the bottom of the panel (All). C Volcano plot for the effect of instrument SNPs on S100B levels shows a symmetric pattern about the zero line.
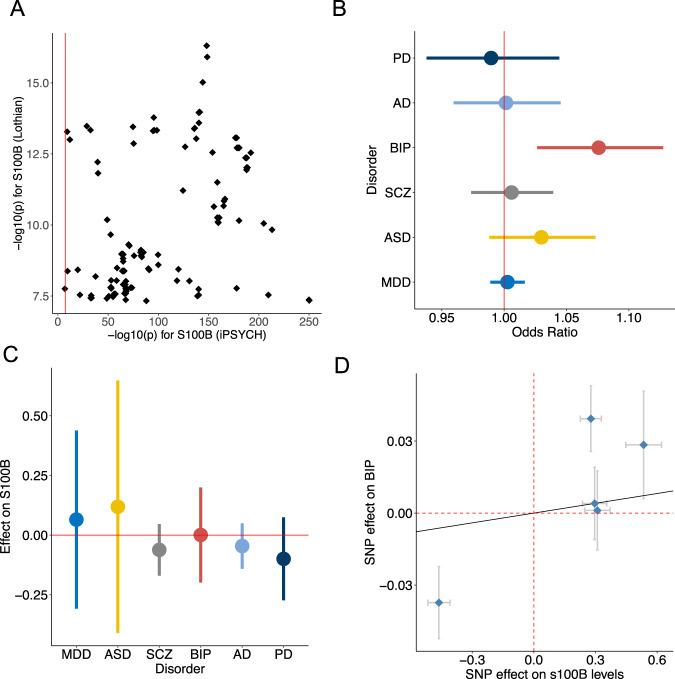
We next compared the association results for S100B levels measured 5-7 days after birth in dried blood spots (the iPSYCH data) to that measured in an older adults sample (the Lothian data) [30]. In total, 156 SNPs existing in both studies were significant in the Lothian data (p < 5 × 10−8), all of which were also significantly associated with S100B in the iPSYCH data (Fig. 3A). However, the association strengths were much stronger in the iPSYCH data than in the Lothian data. Moreover, more than half of these SNPs showed the opposite direction of effects between the two studies (Supplementary Table 35). Altogether, this implies while the same genetic variants influence the circulating levels of S100B, their effect directions and strengths vary with age.
Fig. 3. Bidirectional causal relations between S100B (Lothian) and six neuropsychiatric disorders.
A Comparison of association strengths for SNPs in the Lothian and iPSYCH sample. SNPs that have been discovered in the Lothian study (p < 5e-8) are shown. B IVW MR models were used to estimate average causal effect of circulating levels of S100B measured in the Lothian sample on six neuropsychiatric disorders. Odds ratios and 95% confidence intervals are shown. C Causal effect of six neuropsychiatric disorders on S100B levels. Average effect size (beta) and 95% confidence intervals are shown. D Effects of instrumental SNPs on S100B levels and risk of BIP. PD Parkinson’s disease, AD Alzheimer’s disease, BIP bipolar disorder, SCZ Schizophrenia, ASD autism spectral disorders, MDD major depression disorder.
We then applied our protocol of two-sample MR analysis using the Lothian S100B GWAS results as exposure. Due to the small sample size of this GWAS, very few SNPs passed our selection criteria as instrumental SNPs for S100B upon each disorder (<5, Supplementary Tables 25-30). The causal effect of S100B on MDD, discovered above, was not significant in this data. However, we did find a significant causal effect of S100B on the other mood disorder, BIP: a one SD increase in S100B levels increased the odds ratio of BIP by 1.07 (95%CI: 1.03–1.13; p = 0.002; Fig. 3D). This causal relation was also supported by the weighted median and MR-RAPs methods (Supplementary Table 36). In line with the results from the iPSYCH data, no causal relations were found for the other five disorders, nor for the reverse direction of causality in any disorder (Fig. 3C, Supplementary Tables 37 and 38).
Discussion
We present a comprehensive study aimed at identifying the causal effects of circulating S100B levels on six neuropsychiatric or neurological disorders, including neurodevelopmental (ASD and SCZ), neurodegenerative (AD and PD), and mood disorders (BIP and MDD). We used two datasets for S100B, one measured 5-7 days after birth and the other for those aged over 70 years. We show that increased S100B levels after birth may causally raise the lifetime risk of MDD, a relation that did not exist for S100B levels measured after 70 years of age. However, we did find that increased S100B levels after 70 years of age had a causal effect on the risk of BIP—the other mood disorder included in our study. Thus, both datasets pointed to a causal effect of increased blood S100B levels on the lifetime risk of mood disorders, but not on neurodevelopmental or neurodegenerative disorders. Therefore, future mechanistic studies on the effect of S100B on this group of mental disorders are warranted.
Several reasonings may explain the different findings from the two S100B datasets. Foremost, S100B levels measured 5–7 days after birth may be under the stronger genetic influence than that measured later in life, as reflected by an extremely high SNP-based heritability (h2 = 0.7) in our recent iPSYCH-based GWAS results [29]. Therefore, using SNP instruments will suffer less from unknown environmental confounding on the causal relations between S100B and neuropsychiatric or neurological disorders. Second, it is known that blood levels of S100B could be altered by injury to the brain, medication, and other conditions [22, 23, 42]. Under these conditions, the effect of genetic variants on S100B levels may be different than right after birth. This hypothesis is supported by the comparison of the two S100B GWAS results, which showed similar associations but varying effects, with the caution of incomparable sample sizes for the two. Last, the small sample for the Lothian GWAS may be underpowered to identify enough instrumental SNPs for S100B levels, as less than five SNPs could be selected given our criteria. Thus, when large samples become available, the effect of S100B on neuropsychiatric or neurological disorders will become clearer for this age group.
S100B is a small protein (10–12 kDa), can pass through the blood-brain barrier, and is primarily produced by astrocytes [43]. While it can also be secreted to a lesser extent by adipocytes and possibly other cells, its decay rate from these sources is higher than from astrocytes [43, 44]. Therefore, blood circulating levels of S100B are most likely to be of astrocyte origin. In the brain, at low intracellular concentrations (e.g., nanomolar level), S100B showed neurotrophic effects in vitro, promoting neuronal differentiation, growth and survival [45–47]. But at increased extracellular concentrations it showed neurotoxic effects. While we still lack a mechanistic understanding for this biological phenomenon, it has been proposed that S100B may bind to receptor for advanced glycation end products (RAGE) and induce apoptosis processes in neurons, astrocytes and other glial cells, thereby increasing levels of harmful reactive species in the local extracellular environment. High extracellular levels for S100B can also activate surrounding microglia and astrocytes to promote an inflammatory cellular stage [44]. Although all these biological processes have been implicated as causal pathways leading to neuropsychiatric or neurological disorders, more confirmative data in vivo is still needed.
Surprisingly, we did not observe any causal effect of S100B on four of the neuropsychiatric or neurological disorders studied. On the one hand, our negative result may suggest previously reported associations were influenced by some unmeasured confounding variables. On the other hand, the true causal effect of S100B on these disorders seems very small, such that larger datasets are needed to delineate the effects. We also cannot rule out the third possibility that S100B may causally affect the risk of neuropsychiatric or neurological disorders during some critical windows in the life course outside of the neonatal and older age-ranges measured here, or that the elevated levels of S100B as measured in several studies of neuropsychiatric or neurological disorders are due to other underlying causes than S100B gene variation.
Strengths and limitations
We used the largest GWAS results to date for circulating S100B levels. For the six neuropsychiatric or neurological disorders, we made a trade-off between sample size and the portion of clinical samples included in the original GWAS. We emphasized the latter for ease of interpretation. In addition, the use of S100B measured 5–7 days after birth facilitated a natural cause-effect interpretation in the temporal aspect for disorders developed later in life, which further strengthens our two-sample MR results. However, in this birth sample, S100B was measured in dried blood spots such that its exact levels were not comparable to those measured in serum or whole blood, the material used in the Lothian sample. For this reason, we interpreted the effect of S100B on the unit of standard deviation (SD = 3043 pg/mL) [36]. Because the GWAS of S100B in the Lothian sample were rank-based normalized before analysis [30], our estimated causal effect could not be transferred to other samples. Therefore, our results should be interpreted as evidence of the existence of such a causal effect, but the effect sizes may not be generalizable.
Conclusion
Our data indicates increased circulating S100B levels may causally associate with the lifetime risk of mood disorders. Hence, our findings point to a potential causal explanation behind previously reported S100B associations with brain disorders, possibly opening avenues for novel treatments of major depression and bipolar disorders.
Supplementary information
Acknowledgements
We thank the computational resources provided by UNINETT Sigma2-the National Infrastructure for High Performance Computing and Data Storage in Norway – with project no. (nn9769k/ns9769k). We thank the International Genomics of Alzheimer’s Project (IGAP) for providing summary results data for these analyses. The investigators within IGAP contributed to the design and implementation of IGAP and/or provided data but did not participate in analysis or writing of this report. IGAP was made possible by the generous participation of the control subjects, the patients, and their families. The i–Select chips was funded by the French National Foundation on Alzheimer’s disease and related disorders. EADI was supported by the LABEX (laboratory of excellence program investment for the future) DISTALZ grant, Inserm, Institut Pasteur de Lille, Université de Lille 2 and the Lille University Hospital. GERAD/PERADES was supported by the Medical Research Council (Grant n° 503480), Alzheimer’s Research UK (Grant n° 503176), the Wellcome Trust (Grant n° 082604/2/07/Z) and German Federal Ministry of Education and Research (BMBF): Competence Network Dementia (CND) grant n° 01GI0102, 01GI0711, 01GI0420. CHARGE was partly supported by the NIH/NIA grant R01 AG033193 and the NIA AG081220 and AGES contract N01–AG–12100, the NHLBI grant R01 HL105756, the Icelandic Heart Association, and the Erasmus Medical Center and Erasmus University. ADGC was supported by the NIH/NIA grants: U01 AG032984, U24 AG021886, U01 AG016976, and the Alzheimer’s Association grant ADGC–10–196728. This study is supported by Norwegian Research Council (No. 302854), the UiO:Life Science Convergence environment, University of Oslo, Norway (4MENT).
Author contributions
YW, RN and MEB designed this study; RN, TW and MEB contributed dataset; MP, RN and YW performed analysis; MP, MEB, AMF, JMR, TW, KBW and YW interpreted the results; MP, YW and JMR prepared the first draft; All authors approved the final draft.
Data availability
The datasets supporting the conclusions of this article are publicly available at S100B GWAS results: http://www.ipsych.dk Psychiatric disorder GWAS results: https://www.med.unc.edu/pgc/download-results Alzheimer’s disease GWAS results: https://www.niagads.org/igap-rv-summary-stats-kunkle-p-value-data Parkinson’s disease GWAS results: https://drive.google.com/drive/folders/10bGj6HfAXgl-JslpI9ZJIL_JIgZyktxn Mendelian randomization software Two-Sample MR: https://mrcieu.github.io/TwoSampleMR/ MR-RAPS: https://github.com/qingyuanzhao/mr.raps/tree/multivariate MR-PRESSO: https://github.com/rondolab/MR-PRESSO.
Competing interests
The authors declare no competing interests.
Ethic approval and consent to participate
Each Genome-Wide Association Study received ethic approval from their respective institutional review boards.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41398-023-02478-3.
References
- 1.Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries. 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet (Lond, Engl) 2017;390:1211–59. doi: 10.1016/S0140-6736(17)32154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51:793–803. doi: 10.1038/s41588-019-0397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50:668–81. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet. 2019;51:414–30. doi: 10.1038/s41588-019-0358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat Genet. 2014;46:989–93. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. 2019;51:431–44. doi: 10.1038/s41588-019-0344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schork AJ, Won H, Appadurai V, Nudel R, Gandal M, Delaneau O, et al. A genome-wide association study of shared risk across psychiatric disorders implicates gene regulation during fetal neurodevelopment. Nat Neurosci. 2019;22:353–61. doi: 10.1038/s41593-018-0320-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Dushlaine C, Rossin L, Lee PH, Duncan L, Parikshak NN, Newhouse S, et al. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci. 2015;18:199–209. doi: 10.1038/nn.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Futtrup J, Margolinsky R, Benros ME, Moos T, Routhe LJ, Rungby J, et al. Blood-brain barrier pathology in patients with severe mental disorders: a systematic review and meta-analysis of biomarkers in case-control studies. Brain Behav Immun Health. 2020;6:100102. doi: 10.1016/j.bbih.2020.100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mrak RE, Griffinbc WS. The role of activated astrocytes and of the neurotrophic cytokine S100B in the pathogenesis of Alzheimer’s disease. Neurobiol Aging. 2001;22:915–22. doi: 10.1016/S0197-4580(01)00293-7. [DOI] [PubMed] [Google Scholar]
- 12.Michetti F, D’Ambrosi N, Toesca A, Puglisi MA, Serrano A, Marchese E, et al. The S100B story: from biomarker to active factor in neural injury. J Neurochem. 2019;148:168–87. doi: 10.1111/jnc.14574. [DOI] [PubMed] [Google Scholar]
- 13.Dagdan E, Morris DW, Campbell M, Hill M, Rothermundt M, Kastner F, et al. Functional assessment of a promoter polymorphism in S100B, a putative risk variant for bipolar disorder. Am J Med Genet Part B, Neuropsychiatr Genet. 2011;156b:691–9. doi: 10.1002/ajmg.b.31211. [DOI] [PubMed] [Google Scholar]
- 14.Rothermundt M, Missler U, Arolt V, Peters M, Leadbeater J, Wiesmann M, et al. Increased S100B blood levels in unmedicated and treated schizophrenic patients are correlated with negative symptomatology. Mol Psychiatry. 2001;6:445–9. doi: 10.1038/sj.mp.4000889. [DOI] [PubMed] [Google Scholar]
- 15.Tulner DM, Smith OR, de Jonge P, van Melle JP, Slomp J, Storm H, et al. Circulating cerebral S100B protein is associated with depressive symptoms following myocardial infarction. Neuropsychobiology. 2009;59:87–95. doi: 10.1159/000209860. [DOI] [PubMed] [Google Scholar]
- 16.Matthias LS, Julia S, Johann S, Peter S, Karsten M. Serum S100B Represents a New Biomarker for Mood Disorders. Curr Drug Targets. 2013;14:1237–48. doi: 10.2174/13894501113149990014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomova A, Keményová P, Filčíková D, Szapuová Ž, Kováč A, Babinská K, et al. Plasma levels of glial cell marker S100B in children with autism. Physiol Res. 2019;68:S315–s23. doi: 10.33549/physiolres.934350. [DOI] [PubMed] [Google Scholar]
- 18.da Rosa MI, Simon C, Grande AJ, Barichello T, Oses JP, Quevedo J. Serum S100B in manic bipolar disorder patients: Systematic review and meta-analysis. J Affect Disord. 2016;206:210–5. doi: 10.1016/j.jad.2016.07.030. [DOI] [PubMed] [Google Scholar]
- 19.Gattaz WF, Lara DR, Elkis H, Portela LV, Goncalves CA, Tort AB, et al. Decreased S100-beta protein in schizophrenia: preliminary evidence. Schizoph Res. 2000;43:91–5. doi: 10.1016/S0920-9964(99)00146-2. [DOI] [PubMed] [Google Scholar]
- 20.Yelmo-Cruz S, Morera-Fumero AL, Abreu-González P. S100B and schizophrenia. Psychiatry Clin Neurosci. 2013;67:67–75. doi: 10.1111/pcn.12024. [DOI] [PubMed] [Google Scholar]
- 21.Jang BS, Kim H, Lim SW, Jang KW, Kim DK. Serum S100B Levels and Major Depressive Disorder: Its Characteristics and Role in Antidepressant Response. Psychiatry Investig. 2008;5:193–8. doi: 10.4306/pi.2008.5.3.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houlihan LM, Harris SE, Deary IJ, Starr JM. Replication association analysis of S100B and cognitive ageing. Psychiatr Genet. 2010;20:133–4. doi: 10.1097/YPG.0b013e32833a2096. [DOI] [PubMed] [Google Scholar]
- 23.Schroeter ML, Steiner J, Mueller K. Glial pathology is modified by age in mood disorders-a systematic meta-analysis of serum S100B in vivo studies. J Affect Disord. 2011;134:32–8. doi: 10.1016/j.jad.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 24.Arrais AC, Melo LHMF, Norrara B, Almeida MAB, Freire KF, Melo AMMF, et al. S100B protein: general characteristics and pathophysiological implications in the Central Nervous System. Int J Neurosci. 2022;132:313–21. [DOI] [PubMed]
- 25.Smith GD, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 26.Zhao Q, Wang J, Hemani G, Bowden J, Small DS. Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. Ann Stat. 2020;48:1742–69. doi: 10.1214/19-AOS1866. [DOI] [Google Scholar]
- 27.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–25. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408. doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Nudel R, Benros ME, Skogstrand K, Fishilevich S, i PB, et al. Genome-wide association study identifies 16 genomic regions associated with circulating cytokines at birth. PLOS Genet. 2020;16:e1009163. doi: 10.1371/journal.pgen.1009163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gadd D, McGeachan R, Hillary R, McCartney D, Harris S, Sherwood R, et al. The genetic and epigenetic profile of serum S100β in the Lothian Birth Cohort 1936 and its relationship to Alzheimer’s disease [version 1; peer review: 2 approved] Wellcome Open Res. 2021;6:306. doi: 10.12688/wellcomeopenres.17322.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor AM, Pattie A, Deary IJ. Cohort Profile Update: The Lothian Birth Cohorts of 1921 and 1936. Int J Epidemiol. 2018;47:1042-r. doi: 10.1093/ije/dyy022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howard DM, Adams MJ, Clarke T-K, Hafferty JD, Gibson J, Shirali M, et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22:343–52. doi: 10.1038/s41593-018-0326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nalls MA, Blauwendraat C, Vallerga CL, Heilbron K, Bandres-Ciga S, Chang D, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–102. doi: 10.1016/S1474-4422(19)30320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skrivankova VW, Richmond RC, Woolf BAR, Yarmolinsky J, Davies NM, Swanson SA, et al. Strengthening the Reporting of Observational Studies in Epidemiology Using Mendelian Randomization: The STROBE-MR Statement. JAMA. 2021;326:1614–21. doi: 10.1001/jama.2021.18236. [DOI] [PubMed] [Google Scholar]
- 35.Pedersen CB, Bybjerg-Grauholm J, Pedersen MG, Grove J, Agerbo E, Baekvad-Hansen M, et al. The iPSYCH2012 case-cohort sample: new directions for unravelling genetic and environmental architectures of severe mental disorders. Mol Psychiatry. 2018;23:6–14. [DOI] [PMC free article] [PubMed]
- 36.Skogstrand K, Hagen CM, Borbye-Lorenzen N, Christiansen M, Bybjerg-Grauholm J, Baekvad-Hansen M, et al. Reduced neonatal brain-derived neurotrophic factor is associated with autism spectrum disorders. Transl Psychiatry. 2019;9:252. doi: 10.1038/s41398-019-0587-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ripke S. Ricopili Pipeline And Standards of GWAS Analyses. Eur Neuropsychopharmacol. 2019;29:S713–S4. doi: 10.1016/j.euroneuro.2017.06.020. [DOI] [Google Scholar]
- 38.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kmenta J. Mostly Harmless Econometrics: An Empiricist’s Companion. Bus Econ. 2010;45:75–6. doi: 10.1057/be.2009.37. [DOI] [Google Scholar]
- 40.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693–8. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benjamini Y, Hochberg Y Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Series B. 1995;57:289–300.
- 42.Cox SR, Allerhand M, Ritchie SJ, Muñoz Maniega S, Valdés Hernández M, Harris SE, et al. Longitudinal serum S100β and brain aging in the Lothian Birth Cohort 1936. Neurobiol Aging. 2018;69:274–82. doi: 10.1016/j.neurobiolaging.2018.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Donato R. Functional roles of S100 proteins, calcium-binding proteins of the EF-hand type. Biochim Biophys Acta (BBA) - Mol Cell Res. 1999;1450:191–231. doi: 10.1016/S0167-4889(99)00058-0. [DOI] [PubMed] [Google Scholar]
- 44.Donato R. S100: a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int J Biochem Cell Biol. 2001;33:637–68. doi: 10.1016/S1357-2725(01)00046-2. [DOI] [PubMed] [Google Scholar]
- 45.Kligman D, Marshak DR. Purification and characterization of a neurite extension factor from bovine brain. Proc Natl Acad Sci USA. 1985;82:7136–9. doi: 10.1073/pnas.82.20.7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haglid KG, Yang Q, Hamberger A, Bergman S, Widerberg A, Danielsen N. S-100beta stimulates neurite outgrowth in the rat sciatic nerve grafted with acellular muscle transplants. Brain Res. 1997;753:196–201. doi: 10.1016/S0006-8993(96)01463-1. [DOI] [PubMed] [Google Scholar]
- 47.Winningham-Major F, Staecker JL, Barger SW, Coats S, Van Eldik LJ. Neurite extension and neuronal survival activities of recombinant S100 beta proteins that differ in the content and position of cysteine residues. J Cell Biol. 1989;109:3063–71. doi: 10.1083/jcb.109.6.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the conclusions of this article are publicly available at S100B GWAS results: http://www.ipsych.dk Psychiatric disorder GWAS results: https://www.med.unc.edu/pgc/download-results Alzheimer’s disease GWAS results: https://www.niagads.org/igap-rv-summary-stats-kunkle-p-value-data Parkinson’s disease GWAS results: https://drive.google.com/drive/folders/10bGj6HfAXgl-JslpI9ZJIL_JIgZyktxn Mendelian randomization software Two-Sample MR: https://mrcieu.github.io/TwoSampleMR/ MR-RAPS: https://github.com/qingyuanzhao/mr.raps/tree/multivariate MR-PRESSO: https://github.com/rondolab/MR-PRESSO.