Figure 2.
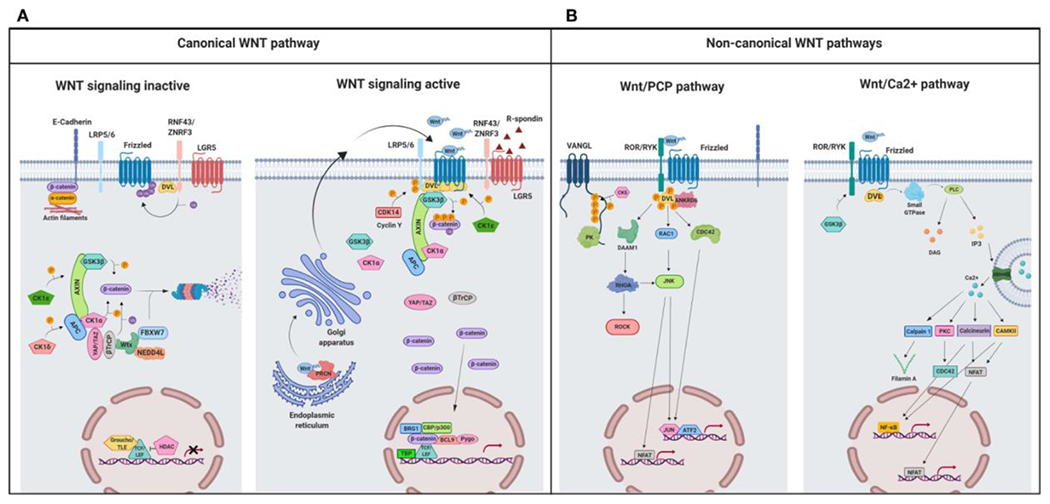
A schematic illustration representing different Wnt signaling pathways. (A) Canonical Wnt signaling. Left panel shows inactive pathway. In the absence of Wnt ligands, β-catenin is phosphorylated by the destruction complex, constituted by the scaffolding proteins APC and AXIN and the kinases GSK3β and CK1α. Then, β-catenin is ubiquitinated and targeted for proteasomal degradation by the complex containing β-TrCP, FBXW7, NEDDL4, and WTX proteins. Thus, β-catenin degradation prevents its presence in the nucleus where a complex formed by TCF/LEF and TLE/Groucho binds HDACs to inhibit transcription of target genes. Right panel shows canonical Wnt signaling active. The binding of Wnt ligands to FZD receptors and LRP co-receptors activates Wnt signaling. LRP receptors are phosphorylated by CK1α and GSK3β. Then, DVL proteins polymerize and are activated at the plasma membrane inhibiting the destruction complex. This results in stabilization and accumulation of β-catenin in the cytosol and its subsequent translocation into the nucleus where it displaces TLE/Groucho repressors forming an active complex with TCF/LEF proteins that bind co-activators such as CBP/p300, BRG1, BCL9, and PYGO. An alternative way of β-catenin signaling includes the disruption of epithelial E-cadherin interactions, which breaks the binding of β-catenin to the cytoplasmic domain of cadherin and leads to the accumulation of β-catenin first in the cytosol, and later in the nucleus. (B) Schematic illustration representing the main non-canonical Wnt pathways. Left panel shows the Wnt/PCP pathway. Wnt ligands bind to the FZD receptor and the co-receptors ROR 1/2 (or RYK). Then, DVL is recruited and activated followed by VANGL activation. Then DVL binds to the small GTPase RHO A with the collaboration of the cytoplasmic protein DAAM1. The small GTPases RAC1 and RHO activate ROCK and JNK. This leads to rearrangements of the cytoskeleton and/or transcriptional responses via for example, ATF2 and/or NFAT. Right panel shows the Wnt/Ca2+ pathway. The signaling is initiated when Wnt ligands bind to the FZD receptor and the co-receptor ROR 1/2 (or RYK). Then, DVL is recruited and activated and binds to the small GTPase which activates phospholipase C leading to intracellular calcium fluxes and downstream calcium dependent cytoskeletal and/or transcriptional responses. APC, adenomatous polyposis coli; BCL9, B-cell CLL/lymphoma 9 protein; β-TrCP, β-Transducin repeat-containing protein; BRG1, Brahma related gene 1; CAMKII, calmodulin-dependent protein kinase II; CBP, CREB-binding protein; CDC42, cell division control protein 42; CELSR, cadherin EGF LAG seven-pass G-type receptor; CK1α,ε,δ, casein kinase 1α,ε,δ; DAAM1, DVL associated activator of morphogenesis; DAG, diacylglycerol; DVL, disheveled; FBXW7, F box/WD repeat-containing protein 7; FZD, Frizzled; GSK3b, glycogen synthase kinase 3β; IP3, inositol 1,4,5 triphosphate; JNK, JUN kinase; LGR5, Leucine-rich repeat-containing G-protein-coupled receptor 5; LRP5/6, low-density lipoprotein receptor-related protein 5/6; NEDD4L, neural precursor cell expressed, developmentally downregulated 4-like; NFAT, nuclear factor of activated T cells; NF-κB, nuclear factor kappa B; PK, Prickle; PKC, protein kinase C; PLC, Phospholipase C; p300, E1A Binding Protein p300; RAC, Ras-related C3 botulinum toxin substrate; RHOA, Ras homolog gene family member A; ROCK, Rho kinase; ROR 1/2, bind tyrosine kinase-like orphan receptor 1 or 2; RYK, receptor-like tyrosine kinase; TBP, TATA-binding protein; PRCN, Porcupine; PYGO, Pygopus; RNF43, Ring finger protein 43; RSPO, R-spondin; TCF/LEF, T-cell factor/lymphoid enhancer factor; TLE, Transducin-Like Enhancer of Split proteins; VANGL, Van Gogh-like; WTX, Wilms tumor suppressor protein complex; YAP/TAZ, Yes-associated protein/Transcriptional co-activator with a PDZ-binding domain; ZNRF3, Zinc and Ring Finger 3. Created with BioRender.com.
From: Martin-Orozco E, Sanchez-Fernandez A, Ortiz-Parra I, Ayala-San Nicolas M. WNT signaling in tumors: the way to evade drugs and immunity. Front Immunol. 2019; 10:2854. doi:10.3389/fimmu.2019.02854.
