Summary
Identifying population-specific genetic variants associated with disease and disease-predisposing traits is important to provide insights into the genetic determinants of health and disease between populations, as well as furthering genomic justice. Various common pan-population polymorphisms at CETP associate with serum lipid profiles and cardiovascular disease. Here, sequencing of CETP identified a missense variant rs1597000001 (p.Pro177Leu) specific to Māori and Pacific people that associates with higher HDL-C and lower LDL-C levels. Each copy of the minor allele associated with higher HDL-C by 0.236 mmol/L and lower LDL-C by 0.133 mmol/L. The rs1597000001 effect on HDL-C is comparable with CETP Mendelian loss-of-function mutations that result in CETP deficiency, consistent with our data, which shows that rs1597000001 lowers CETP activity by 27.9%. This study highlights the potential of population-specific genetic analyses for improving equity in genomics and health outcomes for population groups underrepresented in genomic studies.
Keywords: CETP, Māori, Pacific, Association analyses, HDL-C, Lipids, equity, genetics
Sequencing of CETP identified a common missense variant specific to Polynesian people that associates with higher HDL-C levels and results in CETP deficiency. Given the therapeutic potential of CETP, this study highlights the importance of population-specific genetic analyses of under-represented populations for improving health equity.
Introduction
Dyslipidemia, defined as elevated total or low-density lipoprotein cholesterol (LDL-C) levels, elevated triglycerides, and/or lower high-density lipoprotein cholesterol (HDL-C) levels, is an established risk factor for metabolic diseases such as cardiovascular disease, type 2 diabetes,1 and gout.2 High total cholesterol to HDL-C ratios are prevalent in 40%–44% of Māori and Pacific peoples3 and a higher proportion of Pacific nation ancestry has been associated with lower HDL-C,4,5 consistent with heritability estimates of HDL-C levels (40%–60%).6,7
One locus with genetic variation that consistently associates with lipid levels in multiple population groups, including Pacific peoples, is at the gene that encodes cholesteryl ester transfer protein: CETP.8,9,10,11 CETP modifies lipid levels by mediating the bidirectional transfer of cholesterol esters from atheroprotective HDL-C to atherogenic very-low-density cholesterol in exchange for triglycerides.
In a GWAS for lipids in Samoan people (using samples analyzed in greater depth here), the CETP locus was identified as the most significantly associated locus for HDL-C levels (rs289708, p = 1.19 × 10−11).10 Given this previously reported association with lipid level at CETP in Pacific peoples,10,12 we used a discovery and replication study design13 to investigate whether population-specific genetic variation in CETP could contribute to HDL-C and CETP activity in Māori and Pacific populations.
Here, we describe the identification of a missense variant (reference SNP (rs) cluster ID: rs1597000001 (p.Pro177Leu)) in CETP by sequencing that is specific to Māori and Pacific people. Subsequent association analyses with lipid measures and CETP activity assays show that this variant has very strong effects on HDL levels that are comparable with Mendelian CETP deficiency and drug inhibition of CETP. Genetically lower levels of CETP14,15 have been shown to associate with decreased cardiovascular disease risk and, although CETP inhibitors have not been shown to reduce cardiovascular risk in clinical trials, recent analyses indicate that there might be some cardiovascular16 and metabolic benefits17 to long-term CETP inhibitor use.
While socio-economic inequities contribute to the development and increased impact of metabolic diseases,18,19 research on population-specific genetic variants associated with metabolic traits is important to provide insights into the genetic determinants of phenotypic differences between populations. Ultimately population-specific analyses like the one presented here will address the critical issue of inequity of minority participation in genetic research, furthering genomic justice20 and equity in genomics research for all population groups.
Material and methods
Study cohorts
Demographic and anthropometric characteristics of the participants are summarized in Table 1. A total of 2,272 participants of Māori and Pacific ethnicity were recruited in Aotearoa NZ and served as the discovery cohort in this study. A total 4,309 participants of Samoan ethnicity were recruited in the Independent State of Samoa and the US territory of American Samoa into the Samoan I (n = 2,851), II (n = 908), and III (n = 550) cohorts which served as the replication cohorts. Finally, an additional 255 young people (aged 14–25 years) identifying with at least one Pacific Island ethnicity within Oceania (i.e., Melanesia, Micronesia, or Polynesia) residing in Dunedin, New Zealand, were recruited by the Pacific Trust Otago (PTO) into the PTO Cohort (see supplemental information for additional data). All participants provided written informed consent for the collection of samples and subsequent analysis.
Table 1.
Baseline characteristics of the cohorts
| Aotearoa NZ discovery cohort |
Samoan replication cohorts |
Pacific Trust cohort |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NZ Māori | CI Māori | Samoan | Tongan | Niuēan | Pukapukan | Other/Mixed | Ngāti Porou Hauora | Samoan I | Samoan II | Samoan III | All | |
| Participants (n) | 814 | 172 | 322 | 155 | 37 | 72 | 232 | 270 | 2851 | 908 | 550 | 255 |
| Sex (male) n (%) | 459 (56.39) | 105 (61.05) | 235 (72.98) | 117 (75.48) | 28 (75.68) | 32 (44.44) | 143 (61.64) | 186 (68.89) | 1146 (40.2) | 406 (44.7) | 252 (45.8) | 126 (49.41) |
| Age range | 17–85 | 18–88 | 18–81 | 18–79 | 19–75 | 18–84 | 17–88 | 18–94 | 22–65 | 18–88 | 29–88 | 14–25 |
| Age distribution | 50.92 ± 15.36 | 50.97 ± 15.22 | 44.78 ± 14.18 | 42.17 ± 14.53 | 48.78 ± 13.62 | 44.93 ± 17.2 | 41.34 ± 15.69 | 56.15 ± 13.61 | 45.09 ± 11.18 | 43.26 ± 16.23 | 43.46 ± 8.74 | 18.49 ± 2.54 |
| Height (cm) | 170.31 ± 9.9 | 169.44 ± 10.43 | 173.57 ± 9.05 | 174.48 ± 8.65 | 172.13 ± 8.18 | 165.44 ± 9.57 | 171.74 ± 8.21 | 169.38 ± 8.68 | 165.2 ± 7.8 | 166.6 ± 8.1 | 165.4 ± 7.9 | 172.11 ± 8.95 |
| Weight (kg) | 95.58 ± 23.27 | 100.26 ± 24.53 | 106.04 ± 23.74 | 107.43 ± 20.92 | 98.49 ± 18.59 | 95.6 ± 20.4 | 104.07 ± 28.05 | 100.17 ± 24.81 | 91.3 ± 18.9 | 93.1 ± 22.7 | 87.6 ± 18.3 | 87.98 ± 20.28 |
| LDLa (mmol/L) | 2.74 ± 0.96 | 2.89 ± 0.96 | 2.91 ± 1.03 | 2.85 ± 1.05 | 2.89 ± 0.94 | 2.75 ± 1.1 | 2.78 ± 0.95 | 2.95 ± 1.06 | 3.4 ± 0.86) | 3.2 ± 0.90 | 3.6 ± 0.85 | 2.36 ± 0.64 |
| HDLa (mmol/L) | 1.18 ± 0.4 | 1.18 ± 0.38 | 1.12 ± 0.34 | 1.11 ± 0.37 | 1.1 ± 0.32 | 1.21 ± 0.23 | 1.15 ± 0.37 | 1.15 ± 0.3 | 1.2 ± 0.29) | 1.1 ± 0.26 | 1.0 ± 0.28 | 1.33 ± 0.32 |
| Gout, n (%) | 356 (43.95) | 91 (52.91) | 178 (55.97) | 79 (50.97) | 24 (64.86) | 15 (20.83) | 102 (43.97) | 170 (62.96) | – | – | – | 0 (0.0) |
| Type 2 diabetes, n (%) | 200 (27.47) | 53 (33.97) | 61 (21.79) | 35 (27.13) | 7 (24.14) | 19 (27.14) | 48 (22.64) | 62 (23.66) | 508 (17.8) | 87 (10.1) | – | 0 (0.0) |
The proportions with type 2 diabetes and gout are calculated from individuals without missing data.
Data are mean ± standard deviation or n (%), unless otherwise stated.
NZ, New Zealand; CI, Cook Islands.
Data were unavailable for lipid-lowering medications.
For all participants, information obtained at recruitment included age, sex, height, and weight, as measured by trained assessors. Blood biochemical measurements including lipid measurements were performed at Southern Community Laboratories (Dunedin, NZ) for the Aotearoa NZ and PTO participants, at Northwest Lipid Labs (Seattle, WA, USA) for the Samoan I cohort, and at the Lipids Research Clinic at Miriam Hospital, Brown University for the Samoan II and III cohorts.
The Aotearoa NZ cohort is the amalgamation of three separate groups. A total of 2,002 participants aged ≥16 years, located primarily in Auckland and Christchurch, were recruited to the Genetics of Gout, Diabetes and Kidney Disease in Aotearoa NZ Study.21 The participants from this study were separated into subgroups based on the self-reported Pacific nation ethnicity of their grandparents. Those participants who also reported non-Pacific grandparent ethnicity were grouped according to their Pacific nation ethnicity. This resulted in six Aotearoa NZ sample sets: NZ Māori (n = 814), Cook Island Māori (n = 172), Aotearoa NZ Samoan (n = 322), Tongan (n = 155), Niuēan (n = 37), and an “Other/Mixed” Pacific group (n = 232), which included individuals of Tahitian (n = 1) and Tuvaluan (n = 5) ethnicity, along with individuals who self-reported grandparental ethnicity from more than one Pacific nation (n = 230). An additional 270 participants from Te Tairāwhiti (east coast of the North Island, NZ) were recruited in collaboration with the Ngāti Porou Hauora (Health Service) Charitable Trust. At the request of Ngāti Porou Hauora these participants were analyzed separately to the participants in the six Aotearoa NZ Pacific nation subgroups. Seventy-two participants of Pukapukan ethnicity were recruited in collaboration with the Pukapukan Community of New Zealand in Mangere, South Auckland, NZ.
The Samoan I cohort consists of 2,851 Samoan adults aged 22–65 years residing in the Independent State of Samoa. The Samoan II cohort is a family study consisting of 908 Samoan adults aged 18–88 years residing in the Independent State of Samoa or the US territory of American Samoa. The Samoan III cohort consists of 550 Samoan adults aged 29–88 years residing in the Independent State of Samoa or the US territory of American Samoa. With the exception of participants in the Samoan II cohort, participants were also asked about the ethnicity of each of their grandparents. Participants from the Samoan I and III cohorts all reported four Samoan grandparents. Although participants from the Samoan II cohort did not report the ethnicity of their grandparents, in principal-component analysis of ancestry (see below) the participants of this cohort cluster together with participants from the Samoan III cohort.
Ethical approval for the Aotearoa NZ cohort study was given by the NZ Multi-Region Ethics Committee (MEC/05/10/130; MEC/10/09/092; MEC/11/04/036), the Northern Y Region Health Research Ethics Committee (Ngāti Porou Hauora Charitable Trust study; NTY07/07/074), the University of Otago Human Ethics Committee (PTO; 12/349), and the University of Otago Human Health Ethics Committee (PTO; H17/092). Ethical approval for the Samoan I cohort study was given by the Health Research Committee of the Samoa Ministry of Health and the institutional review board of Brown University. Ethical approvals for the Samoan II and III cohort studies were given by the Health Research Committee of the Samoa Ministry of Health and the institutional review boards of the Department of Health in American Samoan; of The Miriam Hospital, Providence, RI; and of Brown University. The consent forms for Samoan I, II, and III cohorts were available to participants in both Samoan and English. Participants recruited in Aotearoa New Zealand were asked if they would like a karakia (Māori prayer) carried out upon disposal of their blood samples, if indicated this was carried out by the University of Otago ecumenical chaplain. The PTO project was guided by the University of Otago Pacific Research Protocol.
Sequencing of the CETP gene for discovery of Māori and Pacific-specific variants
For genomic sequencing, 2 μg of total genomic DNA from 55 Māori and Pacific individuals were submitted to Kinghorn Center for Clinical Genomics at Garvan Institute of Medical Research in NSW, Australia, for library preparation and next-generation sequencing 30x WGS (TruSeq Nano v.2.5). The CETP gene region (based on reference transcripts from Ensembl) was extracted from whole genome sequence data in FASTQ format aligned to the human genome (GRCh37/hg19) following implementation of the Genome Analysis ToolKit (GATK) best practices using the Burrows-Wheeler Aligner22 (Picardtools) and GATK v.3.6.023 (NeSI pipeline for GATK). Variants in CETP were annotated with allele frequencies from the Genome Aggregation Database24 to identify population-specific variants, and annotated using the Variant Effect Predictor 25 to determine exonic/intronic and synonymous/non-synonymous status. These analyses identified one missense coding variant rs1597000001 (p.Pro177Leu)) in CETP. In silico predictions for deleteriousness of rs1597000001 and conservation at rs1597000001 were obtained using the UCSC browser (GRCh37/hg19) track collection for Combined Annotation Dependent Depletion (CADD) scores and Genomic Evolutionary Rate Profiling (GERP) scores for Mammalian Alignments. The CETP protein structure26 was obtained from The Protein Databank27 and visualized using the PyMOL molecular graphics system (v.2.3.2 (Shrodinger, LLC)).
Genotyping
For participants in the Aotearoa NZ, Ngāti Porou Hauora and PTO cohorts, rs1597000001 was genotyped using a custom-designed TaqMan probe-set (Applied Biosystems, Foster City, CA, USA). A custom Python script (snp_design) was used to annotate the human genome build 37 reference sequence with rs1597000001 and any surrounding SNPs (obtained from the NCBI dbSNP build 147 common SNP list) before primer and probe design. Forward primer: CGGTGCCTGGTACACACTAG; reverse primer: TGTGAACAGCTGCTTGATCCA; probe 1 (VIC): CTGCCTTCAGGCCTG; probe 2 (FAM): CTGCCTTCAGGCTTG. Genotyping was carried out using the LightCycler 480 Real-Time PCR System (Roche Applied Science, Indianapolis, IN, USA) in 384-well plates. For the locus-wide conditional analyses, additional genotypes surrounding rs159700001 at the CETP locus for the Aotearoa NZ participants were obtained from the Illumina Infinium CoreExome v.24 bead chip platform whole-genome genotyping data generated as described previously.21
In the Samoan II and III cohorts, participants were genotyped using the Infinium Global Screening Array-24 v.3.0 BeadChip (Illumina, CA, USA) with custom content that included rs1597000001. A subset of Samoan I cohort participants (n = 1,294) were whole-genome sequenced as part of the Trans-Omics for Precision Medicine (TOPMed) program.28 From this, a Samoan-specific haplotype reference panel was generated from the sequences using Eagle2 (v.2.3.5).29 Using the genotype scaffold and this reference panel rs1597000001 was imputed (R2 = 0.98339) and the additional genotypes for the locus-wide analyses were also imputed in the remaining 1,557 participants in the Samoan I cohort using Minimac4.30
Quality control and quality assurance checks for the Samoan II and III cohorts were conducted using GWASTools.31 The confidence intervals for the minor allele frequency (MAF) of rs1597000001 in the Aotearoa subsets, Samoan cohorts I to III, and Polynesian and non-Polynesian participants of the PTO cohort were computed using the Wilson method and the Agresti-Coull method.32
Generation of principal components and relatedness matrices for use in the association analyses
Whole-genome principal component (PC) vectors were calculated for the Aotearoa NZ cohort using 2,858 ancestry informative markers (as identified by Illumina) extracted from the Infinium CoreExome v.24 whole-genome genotypes. Ten PCs were generated from the SmartPCA (EIGENSOFT v.6.0.1)33 program and used as covariates in the association analyses to account for population stratification and cryptic relatedness. Relatedness coefficients were calculated in the Aotearoa NZ dataset using the software GEMMA (v.0.98.4)34 from 257,069 independent SNPs from the CoreExome whole-genome genotyping data.
For the Samoan I, II, and III cohorts, PCs and empirical kinship coefficients were calculated using genotypes from the Genome-Wide Human SNP 6.0 array (Samoan I) and the Infinium Global Screening Array-24 v.3.0 BeadChip (Samoan II and III). In the Samoan I cohort, PCs were calculated as described in Minster et al.35 and the empirical kinship matrix was calculated from 10,000 independent autosomal markers using OpenMendel.35,36,37 In the Samoan II and III cohorts, 55,640 autosomal markers were used to calculate PCs and empirical kinship matrices with PC-AiR and PC-Relate, respectively,38 as per the recommended procedure in the GENESIS R/Bioconductor package.39
Association analyses of rs1597000001 and the CETP locus in Māori and Pacific people with lipid levels
All association analyses described below were carried out using the R v.4.0.2 software.40 For the association analyses in the Aotearoa NZ cohort, a generalized linear mixed model-based Wald test was carried out using GMMAT software (v.1.3.1)41 to test for associations between rs1597000001 and the continuous variables HDL-C and LDL-C. Analyses were adjusted by sex, age, 10 PCs, and relatedness. A linear model was used to test for associations in the PTO cohort between rs1597000001 and the continuous variables HDL-C and LDL-C using the lm() function in R. The PTO analyses were adjusted by sex, age, and self-reported ethnicity group for each grandparent owing to absence of whole-genome ancestry informative markers in this cohort. For the association analyses in the Samoan I cohort, linear mixed-model regression of the phenotypes on rs1597000001 was performed using the lmekin() function of the coxme R package,42 with sex, age, and four PCs as fixed-effect variables and relatedness as a genetic random-effect variable. In the Samoan II and III cohorts, a mixed model association test was carried out using lmekin()to test for associations between rs1597000001 and HDL-C or LDL-C phenotypes with sex, age, first four PCs, and polity (Samoa or American Samoa) as fixed-effect variables and relatedness as a genetic random-effect variable.
Each Pacific population sample set (Aotearoa NZ cohort: Aotearoa NZ Māori, Aotearoa NZ Cook Island Māori, Aotearoa NZ Samoan, Aotearoa NZ Niuēan, Aotearoa NZ Pukapukan, and Aotearoa NZ Other/Mixed Pacific nations subgroups; Ngāti Porou Hauora group; Samoan Cohort I, Samoan Cohort II, and Samoan Cohort III; and Pacific Trust Otago) was analyzed separately, and the effects combined for the Aotearoa NZ cohort and all cohorts (excluding the PTO cohort) in an inverse variance-weighted fixed-effect meta-analysis using the R package meta (v.3.0-2).43 Heterogeneity between sample sets was assessed using Cochran’s heterogeneity (Q) statistic. The proportion of variance explained by rs1597000001 and the CETP promoter common variant rs1800775 for HDL-C was calculated using the rsq.partial() function in the rsq R package (v.2.2).44 The rsq.partial() function calculates the partial R2 value for each predictor separately including the variants and the covariates sex, age, and PCs, or grandparental ethnicity from the linear model generated by the lm() function in R. The β coefficient in all analyses represents the estimated effect on HDL-C and LDL-C units (mmol/L) per copy of the rs1597000001 T-allele.
To contextualize the effect of rs1597000001 among the other variations in CETP, variants on the Illumina Infinium CoreExome v.24 bead chip platform and present in the Aotearoa NZ cohort (MAF > 0.01) were extracted from the CETP region (rs1597000001 ± 500 kb). A linear model was used to test variants for association with HDL-C using PLINK (v.1.90b6.10)45 adjusted by age, sex, and 10 PCs. Linkage disequilibrium and conditional analyses for rs1597000001 and CETP promoter common polymorphism rs1800775 in the Aotearoa NZ cohort were carried out in PLINK (v.1.90b6.10), adjusting by age, sex, and 10 PCs calculated from the Aotearoa NZ dataset. Conditional analysis was conducted in Samoan cohort I across the CETP region with rs1597000001 modeled as an additional fixed-effect covariate. Regional association plots were created using a custom R package (LocusZoom-like Plots) for the Aotearoa NZ cohort and using LocusZoom46 for the Samoan I cohort.
CETP activity
For the CETP activity assays, individuals of the PTO cohort were recalled specifically for the purpose of providing fresh plasma samples for the CETP activity assays. Fresh plasma (1 μL) was used for the assays, which were carried out in technical triplicate. CETP activity in 11 participants, of whom four were heterozygous for rs1597000001 and seven were homozygous for the rs1597000001 C-allele, was assessed using the CETP activity assay kit II (F) from BioVision (K595-100) according to the manufacturer’s instructions. Fluorescence was measured using a BMG LabTech CLARIOstar plate reader and CLARIOstar software (v.5.01 R2, Firmware 1.10). Statistical analyses were conducted in R v.4.0.2 software with all data reported as means ± standard error of the mean. We used the stat.desc() function of the pastecs R package,47 we tested whether data within each genotypic group were normally distributed. We found that both genotypic groups had skewness/2SE and kurtosis/2SE values between −1 and 1, indicating normality of the data. The Shapiro-Wilks tests for skewness were also non-significant. A two-sample t test with Welch’s correction was conducted to test for a difference in mean CETP activity between the two genotypic groups. One technical replicate was removed after it was identified as an outlier based on a significant Grubbs test for one outlier (p = 0.00065) using the grubbs.test() function from the outliers R package.48
Results
We extracted the coding region of CETP (∼22 kb) from whole-genome sequence data for 55 individuals of Māori and Pacific ethnicity. In our search, we looked for coding variants that have a MAF >1% in the 55 Māori and Pacific discovery genomes but rare (MAF <0.01%) in GnomAD.24 We identified one missense variant in exon 6 of the CETP gene (rs1597000001, CETP:c.530C>T, p.Pro177Leu). The MAF of rs1597000001 was 3.4% (4 heterozygous carriers) in the 55 discovery genomes. The rs1597000001 T-allele was only observed at 0.0036% in the Latino/Admixed American population group in GnomAD,24 which is likely reflects the inclusion of people with Polynesian ancestry in this population group. Our data indicate that rs1597000001 is specific to people of Pacific ethnicity (Figure S1).
rs1597000001 was genotyped in a discovery cohort of 2,270 Māori and Pacific individuals (Table S1) living in Aotearoa New Zealand (NZ). The MAF in the discovery cohort was similar to the 55 Māori and Pacific genomes (3.4%) ranging from 3.2% to 5.4% in the six Aotearoa NZ Pacific nation subgroups (NZ Māori, Cook Island Māori, Aotearoa NZ Samoan, Tongan, Niuēan, and Other/Mixed Pacific) and 2.4% in the Ngāti Porou Hauora group (Table 2). rs1597000001 was monomorphic for the C-allele in the Aotearoa NZ Pukapukan subgroup (MAF = 0.00%, 95% CI; 0.00, 3.12) (Table 2; Figure S1).
Table 2.
Minor allele frequency and Hardy-Weinberg equilibrium of rs1597000001
| Aotearoa NZ Discovery cohort |
Samoan Replication cohorts |
Pacific Trust Otago Cohort |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NZ Māori | CI Māori | Samoan | Tongan | Niuēan | Pukapukan | Other/Mixed | Ngāti Porou Hauora | Samoan I | Samoan II | Samoan III | Polynesian | Non-Polynesian Pacific peoples | |
| Total (n) | 814 | 172 | 322 | 155 | 37 | 72 | 232 | 270 | 2851 | 908 | 550 | 220 | 35 |
| CC (n) (%) |
760 (93.4) | 159 (92.4) | 297 (92.2) | 143 (92.3) | 33 (89.2) | 72 (100.0) | 212 (91.4) | 257 (95.2) | 2600 (91.2) | 825 (90.9) | 497 (90.4) | 201 (91.4) | 35 (100.0) |
| CT (n) (%) |
52 (6.4) | 13 (7.6) | 24 (7.5) | 11 (7.1) | 4 (10.8) | 0 (0.0) | 19 (8.2) | 13 (4.8) | 242 (8.5) | 80 (8.8) | 53 (9.6) | 18 (8.2) | 0 (0.0) |
| TT (n) (%) |
2 (0.3) | 0 (0.0) | 1 (0.3) | 1 (0.7) | 0 (0.0) | 0 (0.0) | 1 (0.4) | 0 (0.0) | 9 (0.3) | 3 (0.3) | 0 (0.0) | 1 (0.5) | 0 (0.0) |
| MAF | 0.034 | 0.039 | 0.040 | 0.042 | 0.054 | 0.000 | 0.045 | 0.024 | 0.046 | 0.047 | 0.048 | 0.045 | 0.000 |
| HWE p | 0.244 | 1.000 | 0.409 | 0.231 | 1.000 | 1.000 | 0.377 | 1.00 | 0.191 | 0.448 | 0.629 | 0.364 | 1.000 |
MAF, minor allele frequency; HWE, Hardy-Weinberg equilibrium; NZ, New Zealand; CI, Cook Islands).
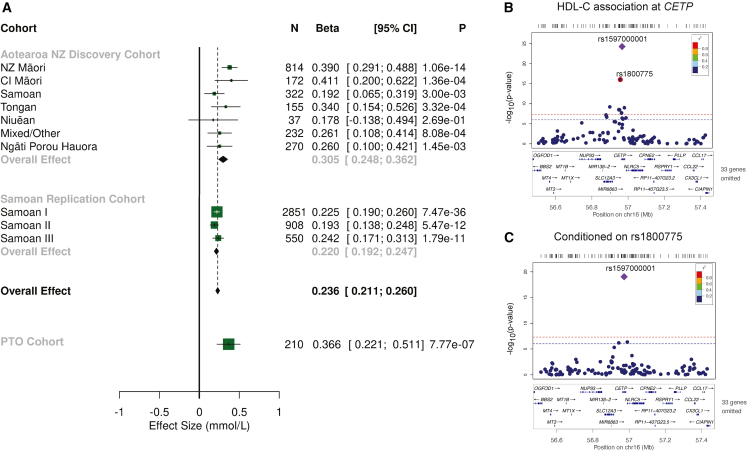
rs1597000001 associated with higher HDL-C levels in those with the rs1597000001 T-allele in all of the surveyed Pacific nation subgroups from Aotearoa NZ (NZ Māori: [95% CI] = 0.390 mmol/L [0.291, 0.488] p = 1.06 × 10−14; Cook Island Māori: = 0.411 mmol/L [0.200, 0.622] p = 1.36 × 10−4; Aotearoa NZ Samoan: = 0.192 mmol/L [0.065; 0.319] p = 3.0 × 10−3; Tongan: = 0.340 mmol/L [0.154; 0.526] p = 3.32 × 10−4; Other/Mixed Pacific: = 0.261 mmol/L [0.108; 0.414] p = 8.08 × 10−4; and the Ngāti Porou Hauora group: = 0.260 mmol/L [0.100; 0.421] p = 1.45 × 10−3), with the exception of the Niuēan subgroup ( = 0.178 mmol/L [−0.138; 0.494] p = 2.69 × 10−1) (Figure 1A). A fixed-effect meta-analysis in the Aotearoa NZ cohort showed a significant overall association of rs1597000001 T-allele ( = 0.305 mmol/L [0.248; 0.361]) p = 1.06 × 10−25; and TT genotype ( = 0.91, [0.61–1.21] p = 5.8 × 10−9) with higher HDL-C, with no evidence of heterogeneity between the Pacific nation subgroups (p = 0.22) (Figure 1A). The proportion of variance in HDL-C levels explained by rs1597000001 in the Aotearoa NZ cohort was 4.5%, similar to proportion of variance explained by sex (5.0%). There was no association of the rs1597000001 with LDL-C (Figure S3) in any of the Pacific nation subgroups, Ngāti Porou Hauora group, or in a fixed-effect meta-analysis of the Aotearoa NZ cohort ( = −0.046 mmol/L [–0.212; 0.120] p = 0.60).
Figure 1.
Association analyses of rs1597000001 T-allele with HDL-C
Forest plot of a fixed-effect meta-analysis for the association of rs1597000001 T-allele with HDL-C (mmol/L) (A). Associations were adjusted by age, sex, 10 PCs, and relatedness in the Aotearoa NZ cohort and age, sex, first four PCs, and relatedness in the Samoan I-III cohorts, and age, sex and number of Pacific and Māori grandparents in the PTO cohort. Association of HDL-C at the CETP locus (+/− 500 kb the lead variant rs1597000001) (B) and after conditioning on rs1800775 (C) using variants on the Illumina Infinium CoreExome v24 bead chip platform for genotyped participants from the Aotearoa NZ cohort. The strength of LD, as measured by the r2, between each variant and rs1597000001, is represented by the color of each point according to the legend in the top right hand corner. The plot was generated using a custom locus zoom-like R package. HDL-C, high-density lipoprotein cholesterol; NZ Māori, New Zealand Māori; CI Māori, Cook Island Māori; PC, principal component; PTO, Pacific Trust Otago; CETP, cholesteryl ester transfer protein.
To replicate the HDL-C association observed in the discovery cohort, we carried out genotyping and association analyses in three independent Samoan cohorts (Samoan I to III) and a fourth cohort consisting of young Pacific people without metabolic disease (PTO cohort). The MAF was 4.4% in Samoan I, 4.7% in Samoan II, and 4.8% in Samoan III (compared with 4.0% in the Aotearoa NZ Samoan cohort) and did not deviate from Hardy-Weinberg equilibrium (HWE) (p > 0.05, Samoan I to III) (Table 2). Association analyses confirmed the association between HDL-C and rs1597000001 (Samoan cohort I: = 0.225 mmol/L [0.190; 0.260] p = 7.47 × 10−36; Samoan cohort II: = 0.193 mmol/L [0.138; 0.248] p = 5.47 × 10−12; and Samoan cohort III: = 0.242 mmol/L [0.171; 0.313] p = 1.79 × 10−11) (Figure 1A), with no evidence of heterogeneity between the sample sets (p = 0.502). The proportion of variance in HDL-C explained by rs1597000001 in the Samoan I, Samoan II, and Samoan III cohorts was 5.1%, 6.4%, and 7.8% respectively. Unlike the Aotearoa NZ cohort, there was an association of the rs1597000001 T-allele with lower LDL-C in Samoan cohorts I and II (Samoan cohort I: = −0.134 mmol/L [−0.238; −0.030] p = 1.15 × 10−2; Samoan cohort II: = −0.276 mmol/L [−0.462; −0.090] p = 3.67 × 10−3), but not Samoan III (Samoan cohort III: = −0.077 mmol/L [−0.312; 0.158] p = 5.21 × 10−1) (Figure S3). In the PTO cohort, rs1597000001 was monomorphic for the major C-allele in Pacific participants without Polynesian ethnicity (i.e., of Melanesian and/or Micronesian ethnicity); however, it is difficult to draw conclusions on this result given the confidence intervals overlap all the other sample sets (MAF = 0.00 [0.00, 6.23]) (Table 2; Figure S1). In PTO participants with Polynesian ethnicity the MAF was 4.5% and did not deviate from HWE (p > 0.05) (Table 2). Association analysis (adjusted by age, sex, and grandparental ethnicity) in PTO participants of Polynesian ethnicity indicated that the rs1597000001 T-allele associates with higher HDL-C levels ( = 0.366 mmol/L [0.221; 0.511] p = 7.77 × 10−7) (Figure 1A) and lower LDL-C levels ( = −0.340 mmol/L [−0.663; −0.016] p = 3.97 × 10−2) (Figure S3). The proportion of variance of HDL-C explained by rs1597000001 in the PTO cohort was 11.8%.
The mean concentration of HDL-C and LDL-C was significantly different in the PTO cohort compared with all Pacific nation subgroups and the Ngāti Porou Hauora group in the Aotearoa NZ cohort (Table S1; Figure S2) (post-hoc Tukey test p < 0.05; all datasets). This likely reflects the different health status of this younger cohort and on this basis the PTO cohort was excluded from subsequent meta-analyses. The meta-analyses of the Aotearoa NZ Pacific nation subgroups, Ngāti Porou Hauora group, and Samoan I to III cohorts demonstrated a strong association of the rs1597000001 T-allele with HDL-C levels ( HDLmeta = 0.236 mmol/L [0.211; 0.260] p = 3.33 × 10−78) with no heterogeneity between the cohorts (p = 0.054) (Figure 1A). The rs1597000001 T-allele associated with lower LDL-C levels in the fixed-effect meta-analysis of the Aotearoa NZ and Samoan cohorts I to III ( LDLmeta = −0.133 mmol/L [−0.209; −0.058] p = 5.90 × 10−4) (Figure S3) with no heterogeneity between the cohorts (p = 0.534).
Using locus-wide genotypes obtained from the Illumina Infinium CoreExome v.24 bead chip platform in the Aotearoa NZ cohort, and from whole-genome sequencing in the Samoan I cohort we re-examined10 the entire CETP locus for association with HDL-C levels in Māori and Pacific peoples (Figures 1B, S4, and S5). rs1597000001, the maximally associated variant at the CETP locus, is in weak linkage disequilibrium with other genetic variants (Aotearoa NZ cohort R2 < 0.2, Figure 1C; and Samoan I cohort R2 < 0.8, Figure S5). The next most significantly associated variant is the CETP promoter polymorphism “−629 C/A” rs1800775 (Figure 1B) in the Aotearoa NZ cohort. The rs1800775 C-allele was present in the Aotearoa NZ cohort (46.0%) at a frequency similar to Europeans (48.0% in gnomAD). The effect size for rs1800775 C-allele ( = −0.090 mmol/L [0.112; −0.069] p = 1.9 × 10−15) was small in comparison with the effect of rs1597000001 ( = 0.301 mmol/L) and the proportion of variance of HDL-C explained by rs1800775 in the Aotearoa NZ cohort was 3.3% compared with 4.5% for rs1597000001. The rs1800775 C-allele associates with the same direction of effect observed previously for Europeans ( = −0.080 mmol/L, p = 3.7 × 10−93)8 and Pacific peoples ( = −0.055 mmol/L, p = 1.7 × 10−4).12
Given the large effect of rs1597000001 on HDL-C and the fact that rs1597000001 and rs1800775 exhibit some linkage disequilibrium (r2 = 0.032) we carried out conditional analyses to test whether the effects at rs1800775 and rs1597000001 were independent of each other. In the Aotearoa NZ cohort the effect on HDL-C persisted for the rs1597000001 T-allele ( = 0.266 mmol/L [0.210; 0.323] p = 5.5 × 10−19) conditioned on the rs1800775 genotype (Figure 1C). When conditioning on rs1597000001, the effect for rs1800775 was attenuated (β = −0.072 mmol/L [–0.094; −0.052] p = 5.6 × 10−11) and rs183130 became the most significantly associated variant at the CETP gene locus ( = 0.087 mmol/L [0.062; 0.112] p = 3.0 × 10−11, r2 with rs1800775 = 0.31) (Figure S4). In the Samoan cohort I conditioning on rs1597000001 resulted in two additional signals marked by rs11076175 (G-allele; = −0.0623 mmol/L [95% CI −0.0876; −0.0369] p = 2.0 × 10−6) and rs4783961 (A-allele; = 0.0480 mmol/L [95% CI 0.0279; 0.0681] p = 3.8 × 10−6) (Figure S5). rs4783961 exhibited modest linkage disequilibrium with rs1800775 (r2 = 0.227) and rs183130 (r2 = 0.623). However, rs11076175 had only weak linkage disequilibrium with rs1800775 (r2 = 0.114) and rs183130 (r2 = 0.014). These data indicate that there are at least two, perhaps three, independent genetic effects at the CETP locus that contribute to HDL-C levels in Māori and Pacific people.
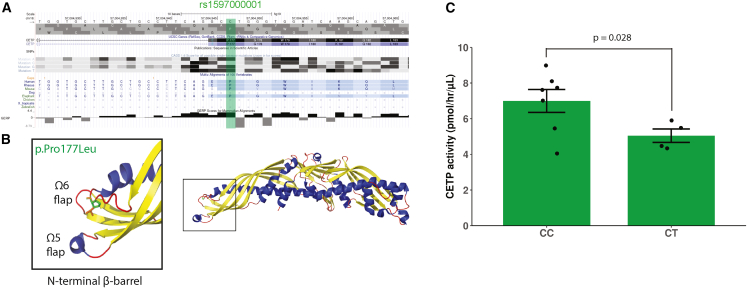
A CADD score of 24.4 places rs1597000001 in the top 1% of predicted deleteriousness, and a GERP score of 4.4 indicated that the variant disrupts an amino acid that is conserved in mammals (Figure 2A). The p.Pro177Leu amino acid substitution corresponds to amino acid position 160 in the mature protein structure of CETP26 owing to the first 17 amino acid residues of the CETP sequence consisting of the signal peptide49 (Figure 2B). To test the hypothesis that rs1597000001 is causal and alters CETP function, we carried out CETP activity assays in serum from participants of the Pacific Trust Otago cohort who were heterozygous for rs1597000001 (n = 4) and homozygous for rs1597000001 C-allele (n = 7). The rs1597000001 T-allele associated with 27.9% lower activity of CETP in comparison with the homozygous carriers of the major C-allele (unpaired t test with Welch’s correction p = 0.028) (Figure 2C) indicating that the rs1597000001 T-allele impacts the function of CETP.
Figure 2.
The population-specific rs1597000001 variant changes the amino acid sequence of CETP and reduces function
(A) The CADD, GERP, and Multiz UCSC track alignment at rs1597000001 for which the substitution creates a non-synonymous amino acid residue change (p.Pro177Leu).
(B) 3D schematic of p.Pro177Leu (P160L) in the mature CETP protein structure in the N-terminal barrel domain (left) of CETP. The CETP protein structure26 was obtained from The Protein Databank27 and illustrated using PyMOL.
(C) Plasma activity of CETP (pmol/h/μL) in 11 participants of the PTO cohort who had the heterozygous rs1597000001 genotype (CT) versus the homozygous rs1597000001 genotype for the major allele (CC). CETP, cholesteryl ester transfer protein. Error bars represent standard error of the mean. A two-sample t test with Welch’s correction was conducted to test for a statistically significant (p = 0.028) difference in mean CETP activity between the two genotypic groups. CETP, cholesteryl ester transfer protein; CADD, Combined Annotation Dependent Depletion scores; GERP, Genomic Evolutionary Rate Profiling.
Discussion
Using whole-genome sequence data from individuals of Māori and Pacific ethnicity, we have identified a population-specific missense variant in CETP that strongly associates with altered lipid profile (higher HDL-C and lower LDL-C) in a meta-analysis of four independent Pacific cohorts. The positive effect on HDL-C levels of the rs1597000001 T-allele was observed in every group analyzed, with the exception of the Niuēan Pacific nation subgroup; however, the effect size estimate was positive, which is consistent with all the other island nation subgroups in the Aotearoa NZ cohort. Given the likely, but unstudied, nutritional and environmental variations across these groups within Aotearoa NZ and between Aotearoa NZ and the Samoan Islands, it is noteworthy that similar effect sizes were detected.
The effects of the rs1597000001 T-allele on LDL-C are less conclusive. In the Samoan I and II cohorts, an association with lower LDL-C was observed. However, the Aotearoa NZ, Samoan III, and PTO cohorts did not show an association of rs1597000001 with LDL-C levels. The differences could be due to unmeasured heterogeneity in lipid-lowering medication use and other genetic and environmental factors. Our findings that rs1597000001 associates with HDL-C consistently throughout Pacific nation groups but not LDL-C, corroborating data from Mendelian randomization studies of CETP, which indicate that CETP activity is an important causal determinant of HDL-C50 levels but not LDL-C levels.
Variants in CETP that cause loss-of-function and consequently have large effects on HDL-C levels are rare in the general population and are enriched in study populations that have high HDL-C levels.51,52,53 Conversely rs1597000001 (MAF ∼2.4%–5.4%) is a low-frequency variant of large effect in Māori and Pacific peoples, and the allele frequency was stable throughout the Pacific nations surveyed here. It is notable that the variant was not detected in people of Pukapukan ethnicity and Pacific peoples without Polynesian ethnicity. Absence of the rs1597000001 T-allele from the Pukapukan subset could reflect a lower minor allele frequency in people of Pukapuka. Alternatively, rs1597000001 is truly absent from the total Pukapukan population, reflecting a different population history for this group, consistent with the oral history of Pukapukan people. That we did not observe the variant in non-Polynesian Pacific people suggests that this is Polynesian specific.
The large effect on HDL-C level that we observe for the rs1597000001 T-allele is reminiscent of that observed for the low-frequency hyperalphalipoproteinemia 1 (elevated HDL-C levels) loss-of-function variant rs2303790 (p.Asp259Gly; HDL = 0.44 mmol/L)54 located in exon 15 of CETP. Functionally, rs2303790 reduces CETP activity, likely emulating Anacetrapib inhibition of CETP.55 Anacetrapib on average raises HDL-C levels by 1.12 mmol/L in patients with atherosclerotic vascular disease.56 Given that rs1597000001 is located in exon 6, it is likely that its functional effect on CETP is disparate to rs2303790. However, it is notable that the effect of the homozygous TT genotype of rs1597000001 is ∼80% of that observed for Anacetrapib.
The p.Pro177Leu located in the N-terminal β-barrel domain of CETP is predicted to penetrate the HDL-C surface,57 and is a putative entry site for cholesterol ester. p.Pro177Leu is contained in the Ω6 flap (Gln155 to Trp162) which separates from the Ω5 flap upon HDL-C penetration, opening the N-terminal distal end for uptake of cholesterol ester.57 Binding of CETP to the HDL-C substrate occurs via hydrophobic interactions,58 and thus the p.Pro177Leu substitution from proline to the hydrophobic amino acid leucine could alter substrate binding and therefore cholesterol ester transfer activity of CETP. Alternatively, proline residues commonly form bends in protein structures,59 and thus the substitution to leucine could disrupt the conformation of the CETP N-terminal domain. Here, we show that heterozygous carriers of rs1597000001 have lower CETP activity, which supports our hypothesis that the rs1597000001 T-allele results in dysfunctional CETP and consequently increases HDL-C. A limitation of our study here was that we were unable to assess the level of CETP deficiency in homozygous carriers owing to their scarcity and inability to recall and therefore we cannot draw conclusions on whether this variant is a complete loss of function variant.
Our study has identified a functional population-specific variant that strongly associates with an altered lipid profile (higher HDL-C and lower LDL-C). This finding was unexpected given the prevalence of dyslipidaemia3 in Maori and Pacific peoples; however, this contradictory result is likely a reflection of the polygenicity of HDL levels and here we do not explore environmental contribution to HDL levels in these populations. There are conflicting reports on whether CETP deficiency has cardioprotective effects. In the study presented here we were unable to report on the effect this variant has on cardiovascular outcomes since there were no reliable cardiovascular phenotype data and therefore it remains to be seen whether this variant also associates with lower cardiovascular event risk. Exploring cardiovascular disease event data for these cohorts in the future will be important for understanding the genetic contribution of rs1597000001 to cardiovascular disease in Māori and Pacific populations. In addition, although the variant has a tangible biological mechanism of effect resulting in CETP deficiency, understanding how this variant (1) alters CETP activity in the homozygous genotype and (2) is modified by other common, smaller-effect variants within the locus will be important information for more effective targeted interventions with pharmaceuticals.60
Importantly, this variant and the accumulating evidence of the presence of other population-specific trait-associated variants21,61,62,63,64,65 emphasizes the significance of population-specific variation and its influence on disease traits. Comprehensive evaluations of genome-wide genetic variation in Māori and Pacific populations are long overdue,66 essential for improvement in health outcomes,19 and critical for equity in genomics and healthcare as we move toward an era of personalized medicine grounded in genetics.
Data and code availability
All software used in the analyses were open source and described in the material and methods. Code written for the analyses are available in GitHub. The data from the Aotearoa NZ and PTO cohorts are not publicly available owing to consent restrictions but can be requested from the corresponding author under an appropriate arrangement. Samoan cohort I data are available from dbGaP (accession no. phs000914.v1.p1). Samoan II and III cohort data (recruited in 2002–2003 and 1990–1995, respectively) are not available as participants had not consented for data sharing.
Acknowledgments
The authors sincerely thank the participants for generously donating their time and information to this study. We sincerely thank the Pukapuka Community Group (Tamaki Makarau), Pacific Trust Otago (Ōtepoti), and Ngati Porou Hauora Charitable Trust (Tairāwhiti), and the communities represented by the Aotearoa NZ cohort and Samoan cohorts, for their partnership in this work. The authors would like to thank J. Drake (Department of Rheumatology, Canterbury District Health Board, Christchurch, New Zealand), J. de Kwant, R. Laurence, C. Franklin, and M. House (all Department of Medicine, University of Auckland, Auckland, New Zealand), J. Harré Hindmarsh, N. Aupouri, R. Akuhata, and C. Ford (all of Ngāti Porou Hauora Charitable Trust, Te Puia Springs, New Zealand), G. Sexton (Counties Manukau District Health Board, Auckland, New Zealand), B. Vincent (Waitemata District Health Board, Auckland, New Zealand), and F. Taungapeau and E. Nabalagi (Pacific Trust Otago) for recruitment. For the Samoan cohorts, we acknowledge the Samoan Ministry of Health, the Samoa Bureau of Statistics, and the American Samoan Department of Health for their support of this research. For additional acknowledgments including funding see supplemental information.
Author contributions
J.M., T.R.M., and M.L. conceived of the study. M.M., R.T., T.J.M., and A.P.-G. coordinated the sampling, genome sequencing, and genome-wide genotyping for the Aotearoa NZ cohort and 55 Aotearoa NZ genome sequences. M. B. and M.C. carried out the genome sequencing data analyses in the 55 genome sequences. For the Samoan I cohort N.L.H. led the field work data collection and phenotype analyses under the supervision of S.T.M. For the Samoan II and III cohorts, S.T.M. led the field work data collection and phenotype analyses. G.S. and H.C. performed genotyping experiments for the Samoan I cohort, and H.C. prepared DNA for genotyping at the Center for Inherited Disease Research for the Samoan II and III cohorts and for sequencing by the TOPMed Program for the sequenced subset of the Samoan I cohort, under the supervision of R.D. J.Z.Z. and J.C.C. performed genotype imputation for the participants in the Samoa I cohort with guidance from R.L.M. and D.E.W. J.M. coordinated the samples and genotyping for the PTO cohort and carried out the genotyping over the Aotearoa NZ cohort. J.M., M.K., and M.L. carried out the association analyses with guidance from T.R.M., D.E.W., and R.L.M. and assistance from N.S., R.T., E.M.R., J.C.C., and T.J.M. M.R. and B.M. carried out the CETP assays supervised by S.M. M.S.R., S.V., and J.T. facilitated fieldwork in Samoa and American Samoa. T.N., M.S.R., S.V., N.H., P.W., F.K., M.T., N.D., L.S., R.M., N.R., and J.d.Z. contributed to the discussion of the public health and cultural implications of the findings. M.L. and J.M. wrote the manuscript with guidance from T.R.M. and contribution from all co-authors. All co-authors contributed to this work, discussed the results, and critically reviewed and revised the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2023.100204.
Contributor Information
Tony R. Merriman, Email: trmerriman@uabmc.edu.
Megan Leask, Email: megan.leask@otago.ac.nz.
Web resources
Ensembl: https://asia.ensembl.org/index.html;ftp://ftp.ensembl.org/pub/grch37
GATK: https://doi.org/10.5281/zenodo.2564243
Genome Aggregation Database: http://gnomad.broadinstitute.org/
GitHub: https://github.com/MerrimanLab/CETP-Project
LocusZoom: https://doi.org/10.5281/zenodo.5154379
NCBI: ftp://ftp.ncbi.nlm.nih.gov/snp
Picardtools: https://broadinstitute.github.io/picard/
Python script (snp_design): https://doi.org/10.5281/zenodo.56250
The Protein Databank: https://doi.org/10.2210/pdb2OBD/pdb;https://www.rcsb.org/
Variant Effect Predictor: www.ensembl.org/Tools/VEP
Zoom-like R package: https://github.com/Geeketics/LocusZooms
Supplemental information
References
- 1.Kolovou G.D., Anagnostopoulou K.K., Cokkinos D.V. Pathophysiology of dyslipidaemia in the metabolic syndrome. Postgrad. Med. J. 2005;81:358–366. doi: 10.1136/pgmj.2004.025601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rasheed H., Hsu A., Dalbeth N., Stamp L.K., McCormick S., Merriman T.R. The relationship of apolipoprotein B and very low density lipoprotein triglyceride with hyperuricemia and gout. Arthritis Res. Ther. 2014;16:495. doi: 10.1186/s13075-014-0495-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gentles D., Metcalf P., Dyall L., Scragg R., Sundborn G., Schaaf D., Black P.N., Jackson R.T. Serum lipid levels for a multicultural population in Auckland, New Zealand: results from the diabetes heart and health survey (DHAH) 2002-2003. N. Z. Med. J. 2007;120:U2800. [PubMed] [Google Scholar]
- 4.Win Tin S.T., Kenilorea G., Gadabu E., Tasserei J., Colagiuri R. The prevalence of diabetes complications and associated risk factors in Pacific Islands countries. Diabetes Res. Clin. Pract. 2014;103:114–118. doi: 10.1016/j.diabres.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 5.Sun H., Lin M., Russell E.M., Minster R.L., Chan T.F., Dinh B.L., Naseri T., Reupena M.S., Lum-Jones A., Samoan Obesity, Lifestyle, and Genetic Adaptations OLaGA Study Group, et al. The impact of global and local Polynesian genetic ancestry on complex traits in Native Hawaiians. PLoS Genet. 2021;17:e1009273. doi: 10.1371/journal.pgen.1009273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qasim A., Rader D.J. Human genetics of variation in high-density lipoprotein cholesterol. Curr. Atheroscler. Rep. 2006;8:198–205. doi: 10.1007/s11883-006-0074-0. [DOI] [PubMed] [Google Scholar]
- 7.Lusis A.J. Genetics of atherosclerosis. Trends Genet. 2012;28:267–275. doi: 10.1016/j.tig.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ridker P.M., Paré G., Parker A.N., Zee R.Y., Miletich J.P., Chasman D.I. Polymorphism in the CETP gene region, HDL cholesterol, and risk of future myocardial infarction: genomewide analysis among 18 245 initially healthy women from the Women's Genome Health Study. Circ Cardiovasc Genet. 2009;2:26–33. doi: 10.1161/CIRCGENETICS.108.817304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mirmiran P., Esfandiar Z., Hosseini-Esfahani F., Koochakpoor G., Daneshpour M.S., Sedaghati-Khayat B., Azizi F. Genetic variations of cholesteryl ester transfer protein and diet interactions in relation to lipid profiles and coronary heart disease: a systematic review. Nutr. Metab. 2017;14:77. doi: 10.1186/s12986-017-0231-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlson J.C., Weeks D.E., Hawley N.L., Sun G., Cheng H., Naseri T., Reupena M.S., Tuitele J., Deka R., McGarvey S.T., Minster R.L. Genome-wide association studies in Samoans give insight into the genetic architecture of fasting serum lipid levels. J. Hum. Genet. 2021;66:111–121. doi: 10.1038/s10038-020-0816-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kenny E.E., Kim M., Gusev A., Lowe J.K., Salit J., Smith J.G., Kovvali S., Kang H.M., Newton-Cheh C., Daly M.J., et al. Increased power of mixed models facilitates association mapping of 10 loci for metabolic traits in an isolated population. Hum. Mol. Genet. 2011;20:827–839. doi: 10.1093/hmg/ddq510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowe J.K., Maller J.B., Pe'er I., Neale B.M., Salit J., Kenny E.E., Shea J.L., Burkhardt R., Smith J.G., Ji W., et al. Genome-wide association studies in an isolated founder population from the Pacific Island of Kosrae. PLoS Genet. 2009;5:e1000365. doi: 10.1371/journal.pgen.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.NCI-NHGRI Working Group on Replication in Association Studies. Chanock S.J., Manolio T., Boehnke M., Boerwinkle E., Hunter D.J., Thomas G., Hirschhorn J.N., Abecasis G., Altshuler D., et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 14.Cupido A.J., Reeskamp L.F., Hingorani A.D., Finan C., Asselbergs F.W., Hovingh G.K., Schmidt A.F. Joint genetic inhibition of PCSK9 and CETP and the association with coronary artery disease: a factorial mendelian randomization study. JAMA cardiology. 2022;7:955–964. doi: 10.1001/jamacardio.2022.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nordestgaard L.T., Christoffersen M., Lauridsen B.K., Afzal S., Nordestgaard B.G., Frikke-Schmidt R., Tybjærg-Hansen A. Long-term benefits and harms associated with genetic cholesteryl ester transfer protein deficiency in the general population. JAMA Cardiol. 2022;7:55–64. doi: 10.1001/jamacardio.2021.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.HPS3/TIMI55-REVEAL Collaborative Group. Writing Committee. Sammons E., Hopewell J.C., Chen F., Stevens W., Wallendszus K., Valdes-Marquez E., Dayanandan R., Knott C., et al. Long-term safety and efficacy of anacetrapib in patients with atherosclerotic vascular disease. Eur. Heart J. 2022;43:1416–1424. doi: 10.1093/eurheartj/ehab863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dangas K., Navar A.-M., Kastelein J.J. The effect of CETP inhibitors on new-onset diabetes: a systematic review and meta-analysis. European Heart Journal-Cardiovascular Pharmacotherapy. 2022;8:622–632. doi: 10.1093/ehjcvp/pvac025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faatoese A.F., Pitama S.G., Gillies T.W., Robertson P.J., Huria T.M., Tikao-Mason K.N., Doughty R.N., Whalley G.A., Richards A.M., Troughton R.W., et al. Community screening for cardiovascular risk factors and levels of treatment in a rural Maori cohort. Aust. N. Z. J. Public Health. 2011;35:517–523. doi: 10.1111/j.1753-6405.2011.00777.x. [DOI] [PubMed] [Google Scholar]
- 19.Merriman T.R., Wilcox P.L. Cardio-metabolic disease genetic risk factors among Maori and Pacific Island people in Aotearoa New Zealand: current state of knowledge and future directions. Ann. Hum. Biol. 2018;45:202–214. doi: 10.1080/03014460.2018.1461929. [DOI] [PubMed] [Google Scholar]
- 20.Reardon J. University of Chicago Press; 2017. The Postgenomic Condition. [Google Scholar]
- 21.Krishnan M., Major T.J., Topless R.K., Dewes O., Yu L., Thompson J.M.D., McCowan L., de Zoysa J., Stamp L.K., Dalbeth N., et al. Discordant association of the CREBRF rs373863828 A allele with increased BMI and protection from type 2 diabetes in Maori and Pacific (Polynesian) people living in Aotearoa/New Zealand. Diabetologia. 2018;61:1603–1613. doi: 10.1007/s00125-018-4623-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., Cunningham F. The Ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu X., Mistry A., Ammirati M.J., Chrunyk B.A., Clark R.W., Cong Y., Culp J.S., Danley D.E., Freeman T.B., Geoghegan K.F., et al. Crystal structure of cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules. Nat. Struct. Mol. Biol. 2007;14:106–113. doi: 10.1038/nsmb1197. [DOI] [PubMed] [Google Scholar]
- 27.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech Z.A., Torres R., Taliun S.A.G., Corvelo A., Gogarten S.M., Kang H.M., et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loh P.-R., Danecek P., Palamara P.F., Fuchsberger C., A Reshef Y., K Finucane H., Schoenherr S., Forer L., McCarthy S., Abecasis G.R., et al. Reference-based phasing using the haplotype reference consortium panel. Nat. Genet. 2016;48:1443–1448. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M., et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gogarten S.M., Bhangale T., Conomos M.P., Laurie C.A., McHugh C.P., Painter I., Zheng X., Crosslin D.R., Levine D., Lumley T., et al. GWASTools: an R/Bioconductor package for quality control and analysis of genome-wide association studies. Bioinformatics. 2012;28:3329–3331. doi: 10.1093/bioinformatics/bts610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown L.D., Cai T.T., DasGupta A. Interval estimation for a binomial proportion. Stat. Sci. 2001;16:101–133. [Google Scholar]
- 33.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 34.Zhou X., Stephens M. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat. Methods. 2014;11:407–409. doi: 10.1038/nmeth.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minster R.L., Hawley N.L., Su C.T., Sun G., Kershaw E.E., Cheng H., Buhule O.D., Lin J., Reupena M.S., Viali S., et al. A thrifty variant in CREBRF strongly influences body mass index in Samoans. Nat. Genet. 2016;48:1049–1054. doi: 10.1038/ng.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lange K., Papp J.C., Sinsheimer J.S., Sripracha R., Zhou H., Sobel E.M. Mendel: the Swiss army knife of genetic analysis programs. Bioinformatics. 2013;29:1568–1570. doi: 10.1093/bioinformatics/btt187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou H., Sinsheimer J.S., Bates D.M., Chu B.B., German C.A., Ji S.S., Keys K.L., Kim J., Ko S., Mosher G.D., et al. OPENMENDEL: a cooperative programming project for statistical genetics. Hum. Genet. 2020;139:61–71. doi: 10.1007/s00439-019-02001-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conomos M.P., Reiner A.P., Weir B.S., Thornton T.A. Model-free estimation of recent genetic relatedness. Am. J. Hum. Genet. 2016;98:127–148. doi: 10.1016/j.ajhg.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gogarten S.M., Sofer T., Chen H., Yu C., Brody J.A., Thornton T.A., Rice K.M., Conomos M.P. Genetic association testing using the GENESIS R/Bioconductor package. Bioinformatics. 2019;35:5346–5348. doi: 10.1093/bioinformatics/btz567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.R Core Team R. 2013. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 41.Chen H., Szpiro A., Chen W., Brehm J., Celedón J., et al. Wang C., Conomos M., Stilp A., Li Z., Yu R.B. Control for population structure and relatedness for binary traits in genetic association studies via logistic mixed models. Am. J. Hum. Genet. 2016;98:653–666. doi: 10.1016/j.ajhg.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Therneau T.M., Therneau M.T.M. 2015. Package ‘coxme’. R package version 2. [Google Scholar]
- 43.Schwarzer G. meta: an R package for meta-analysis. R. News. 2007;7:40–45. [Google Scholar]
- 44.Zhang D. 2021. Rsq: R-Squared and Related Measures.https://CRAN.R-project.org/package=rsq R package version 2.2. [Google Scholar]
- 45.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grosjean P., Ibanez F., Etienne M., Grosjean M.P. 2018. Package ‘pastecs’. [Google Scholar]
- 48.Komsta L. Processing data for outliers. The Newsletter of the R Project. 2006;6/2:10. [Google Scholar]
- 49.UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blauw L.L., Noordam R., Soidinsalo S., Blauw C.A., Li-Gao R., de Mutsert R., Berbée J.F.P., Wang Y., van Heemst D., Rosendaal F.R., et al. Mendelian randomization reveals unexpected effects of CETP on the lipoprotein profile. Eur. J. Hum. Genet. 2019;27:422–431. doi: 10.1038/s41431-018-0301-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee C.J., Park M.S., Kim M., Ann S.J., Lee J., Park S., Kang S.M., Jang Y., Lee J.H., Lee S.H. CETP, LIPC, and SCARB1 variants in individuals with extremely high high-density lipoprotein-cholesterol levels. Sci. Rep. 2019;9:10915. doi: 10.1038/s41598-019-47456-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oldoni F., Sinke R.J., Kuivenhoven J.A. Mendelian disorders of high-density lipoprotein metabolism. Circ. Res. 2014;114:124–142. doi: 10.1161/CIRCRESAHA.113.300634. [DOI] [PubMed] [Google Scholar]
- 53.Rosenson R.S., Brewer H.B., Jr., Barter P.J., Björkegren J.L.M., Chapman M.J., Gaudet D., Kim D.S., Niesor E., Rye K.A., Sacks F.M., et al. HDL and atherosclerotic cardiovascular disease: genetic insights into complex biology. Nat. Rev. Cardiol. 2018;15:9–19. doi: 10.1038/nrcardio.2017.115. [DOI] [PubMed] [Google Scholar]
- 54.Tang C.S., Zhang H., Cheung C.Y.Y., Xu M., Ho J.C.Y., Zhou W., Cherny S.S., Zhang Y., Holmen O., Au K.W., et al. Exome-wide association analysis reveals novel coding sequence variants associated with lipid traits in Chinese. Nat. Commun. 2015;6:10206. doi: 10.1038/ncomms10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jamalan M., Zeinali M., Ghaffari M.A. A molecular dynamics investigation on the inhibition mechanism of cholesteryl ester transfer protein by Anacetrapib. Med. Chem. Res. 2016;25:62–69. [Google Scholar]
- 56.The HPS3/TIMI55–REVEAL Collaborative Group. Bowman L., Hopewell J.C., Chen F., Wallendszus K., Stevens W., Collins R., Wiviott S.D., Cannon C.P., Braunwald E., et al. Effects of anacetrapib in patients with atherosclerotic vascular disease. N. Engl. J. Med. 2017;377:1217–1227. doi: 10.1056/NEJMoa1706444. [DOI] [PubMed] [Google Scholar]
- 57.Cilpa-Karhu G., Jauhiainen M., Riekkola M.L. Atomistic MD simulation reveals the mechanism by which CETP penetrates into HDL enabling lipid transfer from HDL to CETP. J. Lipid Res. 2015;56:98–108. doi: 10.1194/jlr.M054288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang M., Charles R., Tong H., Zhang L., Patel M., Wang F., Rames M.J., Ren A., Rye K.A., Qiu X., et al. HDL surface lipids mediate CETP binding as revealed by electron microscopy and molecular dynamics simulation. Sci. Rep. 2015;5:8741. doi: 10.1038/srep08741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morgan A.A., Rubenstein E. Proline: the distribution, frequency, positioning, and common functional roles of proline and polyproline sequences in the human proteome. PLoS One. 2013;8:e53785. doi: 10.1371/journal.pone.0053785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Merriman T.R. Application of genetic epidemiology to CETP (cholesteryl ester transfer protein) concentration and risk of cardiovascular disease. Circ. Genom. Precis. Med. 2018;11:e002138. doi: 10.1161/CIRCGEN.118.002138. [DOI] [PubMed] [Google Scholar]
- 61.Ji A., Shaukat A., Takei R., Bixley M., Cadzow M., Topless R.K., Major T.J., Phipps-Green A., Merriman M.E., Hindmarsh J.H., et al. Aotearoa New Zealand Māori and Pacific population-specific gout risk variants: CLNK is a separate risk gene at the SLC2A9 locus. J. Rheumatol. 2021;48:1736–1744. doi: 10.3899/jrheum.201684. [DOI] [PubMed] [Google Scholar]
- 62.Tanner C., Boocock J., Stahl E.A., Dobbyn A., Mandal A.K., Cadzow M., Phipps-Green A.J., Topless R.K., Hindmarsh J.H., Stamp L.K., et al. Population-specific resequencing associates the ATP-binding Cassette subfamily C member 4 gene with gout in New Zealand Māori and pacific men. Arthritis Rheumatol. 2017;69:1461–1469. doi: 10.1002/art.40110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klück V., van Deuren R.C., Cavalli G., Shaukat A., Arts P., Cleophas M.C., Crișan T.O., Tausche A.K., Riches P., Dalbeth N., et al. Rare genetic variants in interleukin-37 link this anti-inflammatory cytokine to the pathogenesis and treatment of gout. Ann. Rheum. Dis. 2020;79:536–544. doi: 10.1136/annrheumdis-2019-216233. [DOI] [PubMed] [Google Scholar]
- 64.Patel S.G., Buchanan C.M., Mulroy E., Simpson M., Reid H.A., Drake K.M., Merriman M.E., Phipps-Green A., Cadzow M., Merriman T.R., et al. Potential PINK1 founder effect in Polynesia causing early-onset Parkinson's disease. Movement Disorders. 2021;36:2199–2200. doi: 10.1002/mds.28665. [DOI] [PubMed] [Google Scholar]
- 65.Wang K., Cadzow M., Bixley M., Leask M.P., Merriman M.E., Yang Q., Li Z., Takei R., Phipps-Green A., Major T.J., et al. A Polynesian-specific copy number variant encompassing the MICA gene associates with gout. Hum. Mol. Genet. 2022;31:3757–3768. doi: 10.1093/hmg/ddac094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Emde A.-K., Phipps-Green A., Cadzow M., Gallagher C.S., Major T.J., Merriman M.E., Topless R.K., Takei R., Dalbeth N., Murphy R., et al. Mid-pass whole genome sequencing enables biomedical genetic studies of diverse populations. BMC Genom. 2021;22:666. doi: 10.1186/s12864-021-07949-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All software used in the analyses were open source and described in the material and methods. Code written for the analyses are available in GitHub. The data from the Aotearoa NZ and PTO cohorts are not publicly available owing to consent restrictions but can be requested from the corresponding author under an appropriate arrangement. Samoan cohort I data are available from dbGaP (accession no. phs000914.v1.p1). Samoan II and III cohort data (recruited in 2002–2003 and 1990–1995, respectively) are not available as participants had not consented for data sharing.