Abstract
Our capacity to measure diverse aspects of human biology has developed rapidly in the past decades, but the rate at which these techniques have generated insights into the biological correlates of psychopathology has lagged far behind. The slow progress is partly due to the poor sensitivity, specificity and replicability of many findings in the literature, which have in turn been attributed to small effect sizes, small sample sizes and inadequate statistical power. A commonly proposed solution is to focus on large, consortia-sized samples. Yet it is abundantly clear that increasing sample sizes will have a limited impact unless a more fundamental issue is addressed: the precision with which target behavioral phenotypes are measured. Here, we discuss challenges, outline several ways forward and provide worked examples to demonstrate key problems and potential solutions. A precision phenotyping approach can enhance the discovery and replicability of associations between biology and psychopathology.
A comprehensive understanding of psychopathology requires a systematic investigation of functioning at multiple levels of analysis, from genes to brain to behavior1,2. The development and widespread use of new technologies—including magnetic resonance imaging (MRI) and inexpensive genetic assays—promised to transform our understanding of psychiatric disorders3 and lead to biomarkers that would enhance diagnosis, treatment and prognosis4. However, increasing technological advances and sophistication in the acquisition and analysis of these data have generally failed to produce consistent research findings with broad and significant clinical relevance to the diagnosis and treatment of mental disorders5. Biology–psychopathology associations are typically small6, often fail to replicate7 and generally lack diagnostic specificity8–10. In short, despite decades of work, thousands of studies and hundreds of millions of research dollars, modern neuroimaging and genetic tools have largely failed to uncover clinically actionable insights into psychopathology11,12.
Modest effects and poor replicability have prompted calls to establish consortia-sized samples to identify reproducible biology–psychopathology associations7, with theoretical and empirical studies indicating that problems of low power and replicability can be addressed with sample sizes ranging from the thousands to tens of thousands6,7. This approach has become standard in molecular genetics and has yielded reliable genetic ‘hits’ for several psychiatric disorders12. Recent analyses suggest a similar approach may be necessary for neuroimaging studies6. Other investigators have focused on improving the validity and accuracy of neuroimaging measures, through the use of sophisticated data acquisition techniques13, improved denoising techniques14 and individually tailored analyses15. Similarly, in genetics, growing interest in moving beyond common genetic variation to study high-effect rare variants mandates an order of magnitude increase in sample size16.
In this Review, we suggest that such attempts will have limited success unless we develop more precise or statistically optimized psychiatric phenotypes (that is, observable characteristics or traits). We begin by briefly summarizing the adverse consequences of phenotypic imprecision for discovering reproducible biology–psychopathology associations and highlight some of the most common types of imprecision. We then provide concrete recommendations for precision phenotyping that will help overcome these challenges. Throughout the Review, we provide worked examples of key concepts, using genetic data obtained at the baseline wave (n = 2,218) and behavioral data obtained from the 2-year follow-up wave (n = 5,820) of the Adolescent Brain Cognitive Development (ABCD) study (behavioral data, release 3.0; genetic data, release 2.0)17. These examples support the conclusion that phenotypic imprecision can thwart the consistent detection of potentially important biology–psychopathology associations. In each case, we describe countermeasures that can be deployed to bolster precision and reliability. Taken together, these strands of psychometric theory and empirical data suggest that the systematic adoption of precision phenotyping has the potential to substantially accelerate efforts to understand the neurogenetic correlates of psychopathology and, ultimately, set the stage for developing more effective clinical tools.
Note that we focus on mental health measures in our manuscript because: (1) the limitations of such measures are rarely discussed in comparison with the extensive literature devoted to improving biological measures; (2) prevalent practices to measure behavior are sub-optimal; and (3) addressing these sub-optimal practices is arguably the most cost-effective and quickest way of improving current methodologies. It also merits comment that, while this Review is centered on psychiatric phenotypes, biological measures are also prone to error and may equally contribute to the problems of weak signal in biology–psychopathology association studies18. Thus, our proposals parallel considerable efforts devoted to improving the validity and accuracy of imaging-derived phenotypes13–15, which is sometimes also called precision phenotyping.
The effect of measurement imprecision on detecting and replicating associations between biology and psychopathology
An important step in understanding and treating psychiatric disorders is the identification of pathophysiological mechanisms. Doing so requires the discovery of robust associations between biology and psychiatric phenotypes, an endeavor that is fundamentally constrained by the validity and reliability of the measured phenotypes. Validity concerns the correspondence between a psychological measure and the construct it is designed to measure. If a psychological measure fails to measure a real entity, or changes in the state of that entity fail to produce systematic variations in the psychological measure, any analyses that rely on the psychological measure will be inaccurate. Reliability refers to the consistency of a measure across items, scales, occasions or raters; and is the inverse of measurement error. Lower reliability (higher error) contributes to noisy estimates and decreased accuracy of rank-ordering of individuals when measuring biology–psychopathology associations19. In fact, reliability imposes an upper limit on the magnitude of linear associations that can be detected (that is, observed biology–psychopathology associations are inversely proportional to measurement reliability), mandating larger and more expensive samples for adequate power and reproducibility20 (Box 1). In sum, adequate validity and reliability are necessary for identifying robust and meaningful biology–psychopathology associations20,21.
Box 1. The relationship between measurement reliability and observed effect size.
The relationship between measurement reliability and the observed effect size20 is pertinent to many fields of research. Here, we discuss the issue in relation to psychiatric phenotypes in the context of associations with neurobiology and/or genetics. Constraints on the precision with which psychological attributes can be measured are captured by true score theory (also known as classical test theory), according to which, variance reflecting a psychological measurement includes a stable component that reflects a person’s ‘true score’ and measurement error82:
| (1) |
Thus, according to true score theory, all psychological measurement incorporates measurement error (that is, ‘error-in-variables model’49), some of which reflects: (1) systematic error attributable to other sources of variance that are not of substantive interest, for example method bias; and (2) random measurement error83. Measurement error attenuates associations between variables49. This bias is intuitively demonstrated with respect to the Pearson coefficient of product-moment correlation (r), which forms the basis of many analyses conducted in the literature on biology–psychopathology associations and can be used as an estimate of effect size. It has been demonstrated that the correlation coefficient, r, which is the sample realization of the population parameter rho (ρ), is always a biased estimate of the true association between two variables, x and y49:
| (2) |
where rox,oy is the observed correlation, rtx,ty is the true correlation, and ryy and rxx are the reliability coefficients for variables x and y.
In most cases, the measurement error will be uncorrelated between the variables, resulting in greater dispersion in the data and an attenuation bias of the correlation coefficient and, by extension, smaller and less accurate effect sizes38,49. Relatedly, the standard error (s.e.) for the correlation coefficient increases as a function of smaller samples, n, and smaller effect sizes, r2, resulting in reduced efficiency of estimation84.
| (3) |
Since the probability value of the correlation coefficient is based on the distribution of Student’s t with n − 2 degrees of freedom , smaller effect sizes, as well as smaller samples, lead to lower statistical power. These issues are especially pertinent to measuring psychopathology phenotypes in biomarker research and, critically, will not be resolved simply by increasing sample sizes38. Assuming sample homogeneity, increased sample sizes will only reduce sampling variability () but not proportionally decrease measurement error. The estimates themselves will remain downwardly biased if measurement error is present. Finally, inasmuch as the resulting sample statistic fails to converge on the correct population parameter, it is less likely to be replicated in subsequent samples21.
It is noteworthy that phenotypic precision is a necessary, but not sufficient, condition for uncovering biology–behavior associations. For example, measurement of human intelligence is psychometrically well developed and yet our understanding of the neurobiology and genetics of intelligence is incomplete. The validity and reliability of psychiatric phenotypes can be compromised by a variety of factors, which we collectively refer to as phenotypic imprecision. In this section, we highlight common and pernicious causes of phenotypic imprecision.
Sampling biases
Different research aims demand specific sampling strategies. For studies seeking to identify biology–psychopathology associations, it is important to have samples that are representative of the population of interest and that maximize statistical power for this research design. Sampling biases, non-representative samples and generalizability issues have been broadly discussed in the literature22, but several specific aspects of sampling bias are particularly relevant to the measurement of psychiatric phenotypes in biological association studies. As a primary example, most psychiatric neuroimaging and genetic research has focused on examining case–control differences defined by traditional diagnostic frameworks, such as the Diagnostic and Statistical Manual for Mental Disorders (DSM-5) and the International Classification of Diseases (ICD-11). These frameworks have questionable reliability and validity23, and likely show a limited correspondence with biological correlates (Box 2). Indeed, there is ample evidence that psychiatric phenotypes are dimensional23, indicating that distinctions between cases and controls based on arbitrary clinical cut-points can artificially reduce statistical power for detecting associations with biological measures; the so-called curse of the clinical cut-off’24 (but see ref. 25). The approach may also complicate attempts to identify at-risk individuals with subclinical/subthreshold symptomatology26 and may result in only a subpopulation of the most severely affected individuals being sampled, leading to problems such as Berkson’s bias and the clinician’s illusion.
Box 2. Limitations of traditional approaches to psychiatric nosology.
Existing diagnostic systems, such as DSM-5 and the ICD-11 have clinical utility, facilitating treatment and communication between mental health professionals and consumers of mental health services85. However, the psychopathological concepts invoked by modern nosology may have a tenuous relationship with biological correlates, undermining our attempts to link measurement of behavioral phenotypes with biomarkers3. The limitations of such nosological schemes for informing our understanding of the biology of mental disorders have long been recognized. Initially developed to capture psychiatric signs and symptoms without detailed consideration of etiology or pathophysiology3, diagnostic criteria have since been reified as reflecting, rather than merely indexing, the natural phenomenology of the proposed disease entities themselves, resulting in a conflation of diagnostic criteria with the proposed underlying disorder86. Philosophically, the field has fallen prey to the question-begging fallacy, in which diagnostic categories are investigated as if they are real entities without first asking whether the categories are valid in the first place.
The limitations of traditional nosologies introduce a substantial source of phenotypic imprecision due to questionable validity. Problematically, current diagnostic systems define mental disorders as polythetic-categorical constructs (that is, diagnoses defined by an established minimum number of criteria, not all of which are required for diagnosis). Prototypical symptoms occurring in pre-specified numbers and combinations are conceptualized as forming discrete taxa, underpinning binary diagnostic decisions. However, it is known that mental disorders have a dimensional rather than a taxonomic structure61, with the frequency and severity of symptoms extending as a continuum from the clinical to the subclinical and into the non-clinical range. A related issue is that individuals are generally diagnosed using hierarchical exclusion rules in diagnostic checklists, by which comorbid conditions may be ruled out based on meeting criteria for another disorder. These factors can lead to artificial ‘prototypical cases’ with elevated symptoms and no comorbidity, as well as distort the covariance structure of the data, which can impact subsequent analyses87. Additionally, focusing on a particular diagnostic category assumes homogeneity of symptoms and mechanisms (the homogeneity assumption—the assumption that different people with the same psychiatric diagnosis are phenotypically similar), but individuals with the same diagnosis may exhibit little to no overlap in symptoms (the heterogeneity problem—the grouping of cases with divergent symptom presentations into the same diagnostic category, or the grouping of symptoms with divergent etiology, pathophysiology, course and/or treatment response)34. Co-morbidity between putatively distinct disorders (that is, the comorbidity problem—psychiatric disorders co-occur in the same individuals more often than would be expected for independent entities, suggesting shared phenomenology and etiology)88, and issues of arbitrary clinical cut-offs and ignoring of the clinical significance of subthreshold symptomatology are well-documented limitations of current psychiatric taxonomies89. These limitations obfuscate the search for the neurobiological correlates of psychiatric symptoms and constitute an impediment to future research in this domain90.
A further complication arises with the recruitment of appropriate control groups. Researchers often exclude controls who endorse past or current DSM-5 or ICD-11 diagnoses or other signs of morbidity, resulting in an unrepresentative ‘super control’ group. When compared with a group of patients meeting a diagnostic threshold, the resulting study design embodies an extreme-groups approach rather than a simple dichotomization of a dimensional variable. Such designs, when applied to the study of dimensional phenomena, are known to confer biased effect estimates27. We acknowledge that traditional approaches to clinical description and diagnosis of mental disorders have clinical utility26. However, in this Review, we explore the application and implications of refined approaches to studying the biological correlates of psychopathology in research rather than clinical contexts. The importance of ethnic and demographic diversity with respect to representativeness, ethnic matching of biological measures and generalizability of predictions of behavior from biology, has also been discussed in the literature28,29. Crucially, some cross-cultural initiatives in population neuroscience and genetics have been developed to meet this need29–31.
Minimal and inconsistent phenotyping
The sheer cost and practical challenges of large-scale recruitment and testing often mean that the time and resources available for psychiatric phenotyping are limited32. Minimal or ‘shallow’ phenotyping, is one of the more commonly encountered causes of phenotypic imprecision in biological studies of psychopathology32. Minimal phenotyping is one-shot assessment using single, and sometimes abbreviated, scales. This will increase the proportion of occasion-specific state variance (error) compared with averaging across two or more occasions, thereby attenuating biology–psychopathology associations. Furthermore, minimal phenotyping may fail to capture important aspects of psychopathology that are associated with biological measures.
Aggregation of data in consortia is further complicated by substantive differences in phenotypic assessment across sites. Numerous scales and questionnaires are available for assessing common psychiatric conditions (for example, depression) and these measures vary greatly in their inclusion and emphasis of symptoms33. Minimal phenotyping exacerbates the heterogeneity problem34, because superficially similar cases—for instance, individuals self-reporting a lifetime history of depression in response to a single self-report probe—likely diverge on important, but unmeasured characteristics, dampening effect sizes and power. For example, it has been demonstrated35 that increasing sample sizes for neuroimaging research of schizophrenia may result in samples that are more heterogeneous, which can lead to lower prediction accuracy in machine learning analyses. This aligns with evidence that people diagnosed with schizophrenia and other disorders often show considerable heterogeneity in biological phenotypes36. Similarly, large clinical cohorts forming the reference samples for genome-wide association studies (GWAS) may also be heterogeneous in terms of clinical phenomenology, which is not revealed by minimal phenotyping37. Thus, despite the advantages of large samples, counterintuitively, increasing sample sizes through consortia-like data pooling may result in decreased, rather than increased, signal-to-noise ratio. Therefore, the quest for ever-larger sample sizes, without consideration of precision phenotyping, is neither efficient nor economical, and will not, on its own, ensure the discovery and replicability of biology–psychopathology associations38.
Phenotypic complexity
The use of raw behavioral scores in simple bivariate correlational (or related) analyses with biological variables assumes a unifactorial and non-hierarchical structure of the target phenotype. However, psychiatric phenotypes often have a multidimensional and hierarchical structure (that is, phenotypic complexity). Collapsing complex, multidimensional psychiatric phenotypes (for example, depression) into unitary scores has the potential to obscure biologically and clinically important sources of variance (for example, anhedonia versus guilt)39. Binary diagnostic labels create similar problems. Apart from multidimensionality, psychiatric phenotypes may also exhibit a complex hierarchical structure40. An example of this hierarchical organization is the Hierarchical Taxonomy of Psychopathology (HiTOP) (Box 3 and Fig. 1). At the top of the hierarchy is the p-factor, a broad transdiagnostic liability to all forms of psychopathology41. Situated below the p-factor are narrower dimensions—internalizing, thought disorders, disinhibited externalizing and antagonistic externalizing—specific to particular domains of psychopathology42. Each of these dimensions, in turn, subsumes still narrower symptom dimensions (for example, fear, distress and substance abuse). Too often, simple summary scores ignore this structure, combining both broad and narrow sources of variance43, leading to attenuation of biology–psychopathology associations.
Box 3. The Hierarchical Taxonomy of Psychopathology.
The Hierarchical Taxonomy of Psychopathology (HiTOP) model is a potentially useful framework for precision psychiatric phenotyping. HiTOP is a data-driven approach to psychiatric nosology that organizes symptoms into homogeneous, hierarchically organized dimensions (Fig. 1)42. The problem of arbitrary diagnostic thresholds, subthreshold/subclinical symptomatology and low power is addressed by measuring psychopathology continuously with no artificial demarcation point designating health from disorder42. The comorbidity problem and heterogeneity problem are addressed by organizing co-occurring problems into homogeneous dimensions42. For example, the high comorbidity of major depressive disorder and generalized anxiety disorder are seen to reflect the operation of common etiological mechanisms, which are captured by the distress subfactor, which is situated under the broader internalizing spectrum within the HiTOP model. Thus, the broadest dimensions, reflecting common liabilities to psychopathology, are situated at the top of the hierarchy with the narrowest traits and symptom components situated at the bottom, reflecting liabilities to specific problems.
The development of an omnibus measure of the HiTOP modelis nearing completion and will be open-source and freely available for use without charge in both computerized and paper-and-pencil formats91. In the meantime, several existing instruments can be used to reliably assess HiTOP dimensions in youth and adults92. HiTOP-conformant measures enable broadband, transdiagnostic assessment of psychopathology at multiple levels of the hierarchy, from broad superspectra dysfunction and spectra to narrower subfactors and empirical syndromes. HiTOP-conformant measures focus on narrow homogneous and unidimensional constructs with high discriminant validity facilitating high reliability and valid inference43,66 for association studies with biology. At the lowest levels of the hierarchy, HiTOP encompasses even narrower symptom components (for example, anhedonia, insomnia) and maladaptive traits42. The latter provides a measure of the lower range and adaptive end of the psychopathology continuum. Combining measures of traits and psychopathology thus improves phenotypic resolution (that is, the reliability or precision of measurement of a phenotype along the full spectrum of the latent trait continuum). Notably, the higher order spectra of the HiTOP model are invariant across sexes and different age groups93. HiTOP dimensions, including the broad superspectra and spectra, as well as narrower subfactors and symptom components, can serve as phenotypic targets for neuroscience-informed Research Domain Criteria (RDoC) domains94.
Fig. 1 |. The HiTOP model.
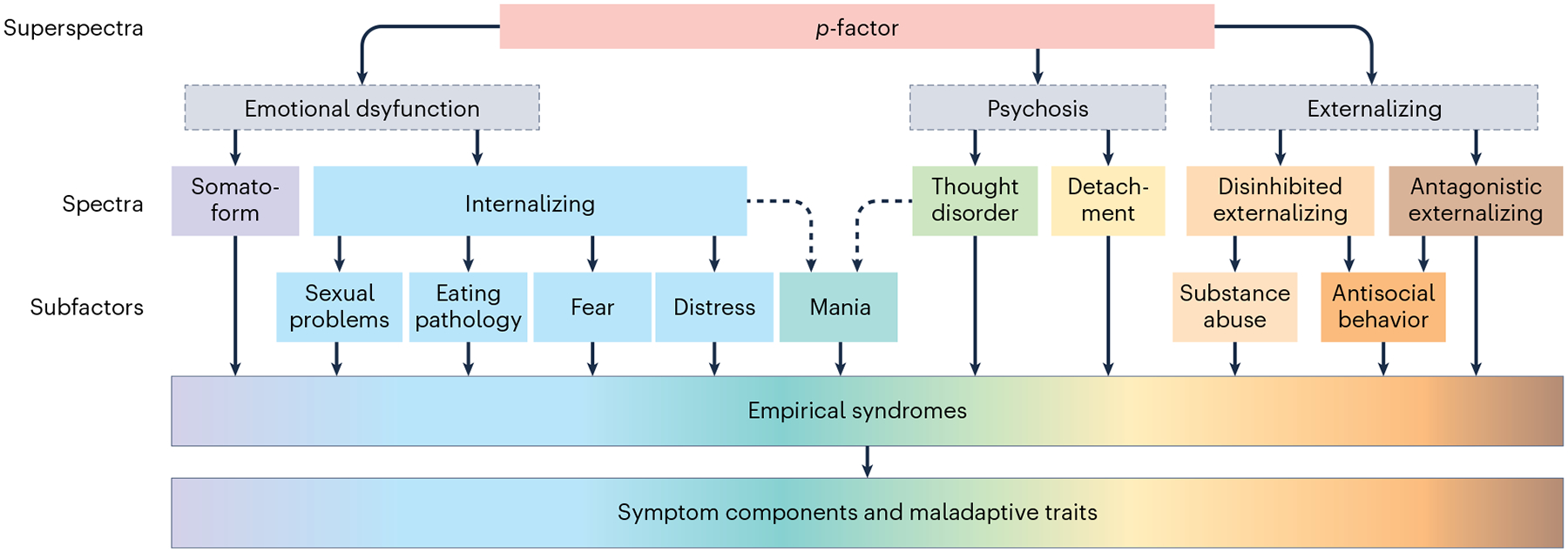
The broadest dimensions, reflecting common liabilities to psychopathology, are situated at the top of the hierarchy with the narrowest traits and symptom components situated at the bottom, reflecting liabilities to specific problems. Gray boxes with broken lines indicate hypothesized, but not yet confirmed, constructs. The broken single-headed arrows pointing to ‘Mania’ reflect preliminary relationships awaiting further confirmatory evidence.
We show in example 1 of the Supplementary Information how failing to differentiate these multidimensional and hierarchical sources of variance from each other can confound relations with biological parameters. We provide an illustration of these concepts using Child Behavior Checklist (CBCL) data from the ABCD study, which exhibits both multidimensionality and hierarchical structure. The CBCL is a multidimensional instrument that measures eight empirical syndromes using eight distinct subscales. The CBCL has a hierarchical structure with variance attributable to three levels: (1) a p-factor; (2) internalizing and externalizing dimensions; and (3) the eight specific psychopathology syndromes. We used a bifactor model44 within a structural equation modeling (SEM) framework (Box 4 and Fig. 2) to separate these dimensions into three orthogonal (that is, uncor-related) variance components and examined how much variance was unique to each level. The CBCL has three composite scales: (1) total problems, which summarizes the scores across the eight syndrome scales; (2) internalizing problems, which summarizes scores across the three internalizing scales; and (3) externalizing problems, which summarizes scores across the two externalizing scales. Less than 49% of the total variance is common across the eight scales, such that collapsing measurement of psychopathology into the unidimensional total problems score misrepresents the data and would result in attenuation of biology–psychopathology associations unique to the p-factor by 30.2% (that is, rxx = 0.488), even assuming perfect reliability of the biological measure. This is despite the total problems score showing high reliability in terms of Cronbach’s alpha (α = 0.949). Thus, it is possible for internal consistency reliability to be high in the presence of multidimensionality, meaning that reliability cannot be used as a unidimensionality statistic.
Box 4. Structural equation modeling.
Hierarchical modeling, measurement invariance, mixture modeling and the T(M-1) model can be done within an SEM framework. SEM is a statistical technique that combines factor analysis, canonical correlation and multiple regression95. SEM can be used to extract the common variance from factor indicators of the construct of interest. The resulting factor, also known as a latent variable, is a purer measure of the construct of interest because only variance common to all variables that reflect the dimension of interest are included as shared variance95. In the common factor model estimated within the SEM framework, reflective latent variables (that is, an underlying factor is conceptualized as causing the covariance in the indicators) are estimated by decomposing observed variables into variance shared with the other factor indicators and variance that is unique to the variable (that is, variance attributable to a separate construct and measurement error). The formula is expressed as:
| (4) |
where xi is a measured variable (that is, observed or manifest variable), ax is an intercept, λx is a factor loading determining the influence of a factor ξi on the measured variable, and θεi is the unique variance or error of the measured variable that is not explained by the factor loading (Fig. 2). This model formalizes the following: (1) the target psychopathology phenotype is unobserved and must be inferred by one or more measured variables (for example, questionnaire items); (2) measured variables are imperfect indices of the target construct and incorporate measurement error; (3) factor indicators are not necessarily equally important measures of the target latent variable, as indicated by differences in the strength of the factor loadings (that is, λx).
In a structural regression model, SEM enables estimation of regression path coefficients between factors within the model. Thus, SEM estimates the empirical relationships between predictor variables and criterion variables with measurement error excluded from the final model95. An additional advantage of using SEM is that hypothesized multiple dependence relationships can be examined concurrently, along with complex interactions95. By contrast, some researchers use a two-step factor score regression technique in which factor scores estimates are derived from the latent variables as manifest variables and then incorporated into subsequent regression analyses. It is important to note that factor score estimates are not the same as latent variables due to factor score indeterminacy. In simple terms, factor score indeterminacy reflects the fact that an infinite set of factor scores can be estimated for the same analysis that will be equally consistent with the factor loadings. This is because the number of observed variables is less than the number of common and unique factors to be estimated96. The degree of factor score indeterminacy is related to the number of factor indicators and their communalities (that is, how much variance is explained in the variables by the factor) and is represented by a validity coefficient, which will vary between studies96. Factor score estimates can, therefore, misrepresent the rank ordering of individuals along the factor96. The degree to which factor score estimates preserve the correlations amongst the factors in the analysis (that is, correlational accuracy) and are not contaminated by variance from orthogonal factors (that is, univocality) will also vary between studies96. The use of factor score estimates can also potentially bias the parameter estimates of the regression models97. Thus, we recommend against this approach in favor of SEM.
Ideally, biological measurements should be incorporated directly into latent models to capitalize on the increased measurement precision and statistical power that these models afford (for example, ref. 98). However, SEM generally requires sample sizes greater than 20099. Thus, it may not be feasible for many research studies examining biological variables. Several SEM packages are commercially available, such as Mplus (http://www.statmodel.com/), and freely available as open-source software, such as lavaan in R (https://lavaan.ugent.be/). The HiTOP Consortium provided a primer for conducting SEM research in the context of dimensional hierarchical models of psychopathology69 and there are several excellent entry-level texts for SEM, such as ref. 99.
Fig. 2 |. The reflective latent variable model.
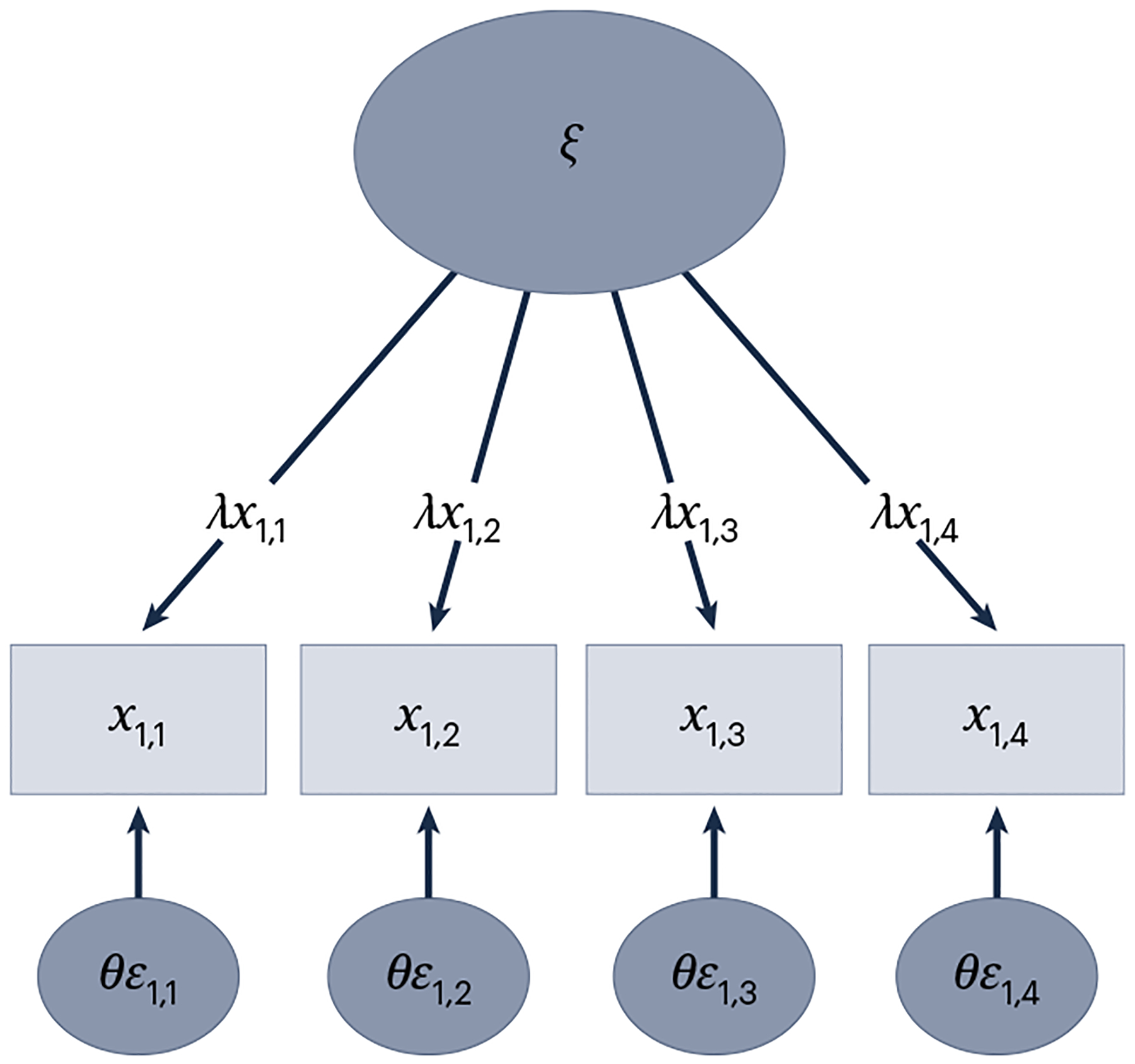
Reflective latent variable (common factor) model in which the unobserved psychobiological attribute (factor or latent construct; ξ), is conceptualized as explaining the variance/covariance in the measured variables (x1,1–x1,4) via their factor loadings (λx1,1–λx1,4), which are linear regression coefficients. The indicator error variances (also residual variances or uniquenesses; θε1,1−θε1,4) capture the variance in each measured variable not explained by the factor (that is, variance not shared with the other indicator variables).
Results are worse for the other two composite scales, internalizing problems and externalizing problems, where variance uniquely attributable to these group dimensions is only 10.4% and 20.1%, resulting in a 67.8% and 55.2% attenuation of correlation coefficients with external variables, respectively (rxx = 0.104 and 0.201). We also demonstrate that high phenotypic complexity across the eight empirical syndrome scales due to the hierarchical organization of the CBCL dimensions leads to low internal consistency reliability for these individual scales (that is, an average of approximately 42% variance is unique to each scale). This low reliability results in substantial attenuation bias, with correlations between symptoms and biological criterion variables being reduced from between 15% (rxx = 0.721 for somatic complaints) to 48.2% (rxx = 0.232 for the anxious/depressed scale).
Inadequate phenotypic resolution
The vast majority of biology–psychopathology association studies implicitly assume that measurement precision is uniform across the latent trait continuum, a concept referred to as phenotypic resolution40. Yet most measured psychiatric phenotypes lack sufficient coverage of the adaptive (low) end of the continuum, leading to differential phenotypic resolution across the range of the scale45. Consider anxiety. Low scores on a clinical scale are meant to represent the absence of pathological anxiety, but often there is little to no item content addressing the opposite end of the latent trait continuum. As a result, there will be high error at the low end of the scale, making it difficult to conduct robust individual differences research. This problem is known as a ‘multiplicative error-in-variable model’, in which the error is proportional to the distributional properties of the signal33. Attenuation bias will thus be present for participants who score at the lower end of the psychopathology continuum, which tends to be most individuals, particularly in studies of community-dwelling, non-clinical populations. The multiplicative error-in-variable model also results in marked heteroscedasticity (that is, the distribution of the residuals or error terms in a regression analyses is unequal across different values of the measured values), which reduces statistical power46.
Phenotypic resolution can be examined using item response theory (IRT; Box 4). IRT provides total information functions, which plot the measurement precision of a phenotype as a function of the standardized latent trait distribution47. Typically, for unipolar psychiatric phenotypes, reliability is unacceptably low (rxx < 0.6) below the mean48. Because reliability places an upper bound on associations with other variables49, this decrease in measurement precision can markedly decrease signal-to-noise ratio in biology–psychopathology association studies.
In example 2 of the Supplementary Information, we provide an illustrative example of poor phenotypic resolution using CBCL data from the ABCD study, with results demonstrating that only a small portion of the sample has reliable scores for most of the CBCL scales. Specifically, we find unacceptably low reliability, even for basic research purposes (rxx < 0.6), at or below one standard deviation below the mean for ten of the eleven scales (that is, all scales except the total problems scale). The average proportion across CBCL scales of the ABCD sample that would not have interpretable scores due to low phenotypic resolution was 37.2% and more than half of the sample had uninterpretable scores for three of the eleven CBCL scales. Thus, despite the promise of the ABCD study for providing a sample size sufficient to accurately assess biology–psychopathology associations, a large proportion of participants from the ABCD study have CBCL scores with unacceptably low reliability, which will have the unfortunate and counterproductive goal of attenuating biology–psychopathology associations.
Measurement non-invariance
Another challenge to the accurate assessment of biology–psychopathology associations is the assumption that a measure assesses a psychiatric construct similarly across groups and measurement occasions (that is, measurement invariance)50. Yet there is ample evidence that measurement properties can vary (that is, non-invariance) across demographic groups (for example, sex) or unobserved or latent classes (that is, homogeneous subpopulations or subgroups, clusters or mixtures, embedded within the sample)51. Non-invariance can substantially bias results, because raw scores do not have the same substantive interpretation across groups. For example, a raw score of 10 on a particular scale may not correspond to the same level of psychopathology in males and females.
Invariance testing provides a rigorous means of evaluating the equivalence of model parameters across groups by imposing a series of increasingly restrictive equality constraints on the model parameter estimates within a factor analytic framework50. Typically, three levels of invariance are evaluated: (1) weak invariance; (2) strong invariance; and (3) strict invariance (Supplementary Table 3 contains technical definitions)50. Unfortunately, only a small proportion of studies test for full measurement invariance50; thus, combining raw scores across discrete groups (for example, sex and ethnicity) for biology–psychopathology associations remains problematic. In example 3 of the Supplementary Information, we provide a striking example of measurement non-invariance of the CBCL total problems scale (which is the most reliable scale of the CBCL)52 between male and female ABCD participants. Results demonstrate that CBCL raw scores are not comparable between male and female children at any point along the latent trait continuum. Thus, any study that pools the results on the CBCL total problems scale for male and female children and tests the association with biological variables will draw erroneous conclusions.
The heterogeneity problem
The heterogeneity problem is increasingly recognized as a key challenge for biological studies of psychiatric illness34. Heterogeneity can be described at person-centered and variable-centered levels34. Person-centered heterogeneity refers to the presence of clusters or subtypes within groups, such as a group of individuals diagnosed with major depression. To the extent that such clusters or subtypes are unrecognized and associated with distinct biological signatures, they will attenuate biology–psychopathology associations (that is, mixing apples and oranges). This problem is exacerbated in case–control research because traditional DSM and ICD diagnoses likely encompass phenomenologically, etiologically and biologically heterogeneous syndromes (Box 2). The result is the so-called ‘jingle fallacy’, in which divergent phenomena are arbitrarily equated, in this case because of the application of a common term53. Variable-centered heterogeneity describes admixtures of symptoms with divergent etiology, pathophysiology, course and/or treatment response54 or a failure to differentiate between narrower homogeneous and unidimensional symptom components.
Both person-centered and variable-centered heterogeneity have emerged as a critical issue in depression research. For example, an analysis of 3,703 participants in a clinical trial for the treatment of depression revealed a remarkable degree of person-centered disorder heterogeneity with 1,030 unique symptom profiles identified using the Quick Inventory of Depressive Symptoms (QIDS-16), 864 (83.9%) of which were endorsed by five or fewer participants and 501 (48.6%) were endorsed by only one participant55. Thus, methodologies that explicitly accommodate potential clinical sample heterogeneity are a promising way forward in psychiatric research56. There is also evidence of variable-centered heterogeneity in depression, which has a clear multifactorial structure despite often being treated as a unitary construct based on sum scores on inventories, such as the Hamilton Rating Scale for Depression57. Indeed, three distinct genetic factors were identified that explained the co-occurrence of distinct subsets of DSM criteria and symptoms: cognitive and psychomotor symptoms, and mood and neurovegetative symptoms58. Heterogeneity has also been identified across depression symptoms in terms of etiology, risk factors and impact on functioning57. These findings suggest that the analysis of narrower homogeneous and unidimensional symptom components or even individual symptoms is likely to be a more informative and productive avenue for future biology–psychopathology association studies.
Method bias
Method bias (sources of systematic measurement error stemming from the measurement process, such as method effects, for constructs) is a common, yet often neglected, potential source of measurement error in biology–psychopathology association studies. Sources of method bias include response styles commonly encountered in self-report, such as social desirability (that is, responses attributable to the desire to appear socially acceptable), acquiescence (‘yea-saying’), disaquiescence (‘nay-saying’), extreme (selecting extreme response categories in Likert-type ordinal scales), and midpoint (selecting middle categories in Likert-type ordinal scales) response styles59. Method bias can distort dimensional structure, obscure true relationships between constructs and compromise validity60,. Method bias is caused by method factors, which describe sources of systematic measurement error that contribute to an individual’s observed score, thus attenuating subsequent analyses of association60. Indeed, method biases are one of the most important sources of measurement error59. Between one-fifth and one-third (18–32%) of the variance in self-report measures is attributable to method factors60. Method factors and the resulting method bias represent serious threats to study validity because, as systematic sources of error variance, they attenuate and otherwise distort the empirical relationship between variables of interest59.
Recommendations for precision psychiatric phenotyping
In this section, we outline some recommendations for enhancing the precision of psychiatric phenotyping and, ultimately, increasing the robustness and reproducibility of biology–psychopathology association studies (Table 1 and Fig. 1).
Table 1 |.
Sources of imprecision in psychopathology phenotyping and proposed solutions
| Problem | Solution |
|---|---|
| Sampling bias | Dimensional sampling and measurement |
| Minimal and inconsistent phenotyping | Deep phenotyping and use of standardized measures |
| Phenotypic complexity | Use of homogeneous unidimensional scales, test for multidimensionality and model hierarchical relations between dimensional constructs |
| Poor phenotypic resolution | Increase phenotypic resolution by adding items assessing the adaptive end of the continuum |
| Measurement non-invariance | Test for and accommodate measurement invariance |
| The heterogeneity problem | |
| Person-centered heterogeneity | Mixture modeling |
| Variable-centered heterogeneity | Broadband assessment of psychopathology and hierarchical modeling |
| Method bias | Multi-method assessment |
Dimensional sampling and measurement
To overcome the limitations of categorical nosological systems, some have advocated for studying dimensional phenotypes that cut across traditional diagnostic categories, a view that closely aligns with the National Institute of Mental Health (NIMH) RDoC2 initiative. Psychometrically, mental disorders show a dimensional rather than a taxonomic structure61 and dimensional measures of psychopathology exhibit greater reliability and validity than categorical diagnoses23. Indeed, the highly polygenic architecture of many psychopathology phenotypes implies that they are dimensionally distributed quantitative traits62. Greater statistical power can be further achieved in biological studies through a dimensional enhancement strategy, involving the recruitment of participants with subthreshold and non-clinical levels of symptoms to leverage symptom variation across the full spectrum of severity63. The chances of sampling bias and clinical heterogeneity will be reduced, and effect size estimates will be less biased, with dimensional (versus case–control study) designs27. Dimensional sampling strategies are potentially more economical than case–control sampling, as dimensional designs do not rely on thorough clinical pre-screening of participants prior to their inclusion in the study64. Dimensional sampling is also more likely to yield samples more representative of the population than case–control sampling, as dimensional sampling does not exclude individuals based on arbitrary clinical cut-offs and hierarchical exclusion rules43. However, to ensure sampling of the full spectrum of symptom or syndrome severity, participants likely to have elevated levels of the target psychopathology dimensions can be over-sampled (Fig. 3).
Fig. 3 |. Precision psychiatric phenotyping.
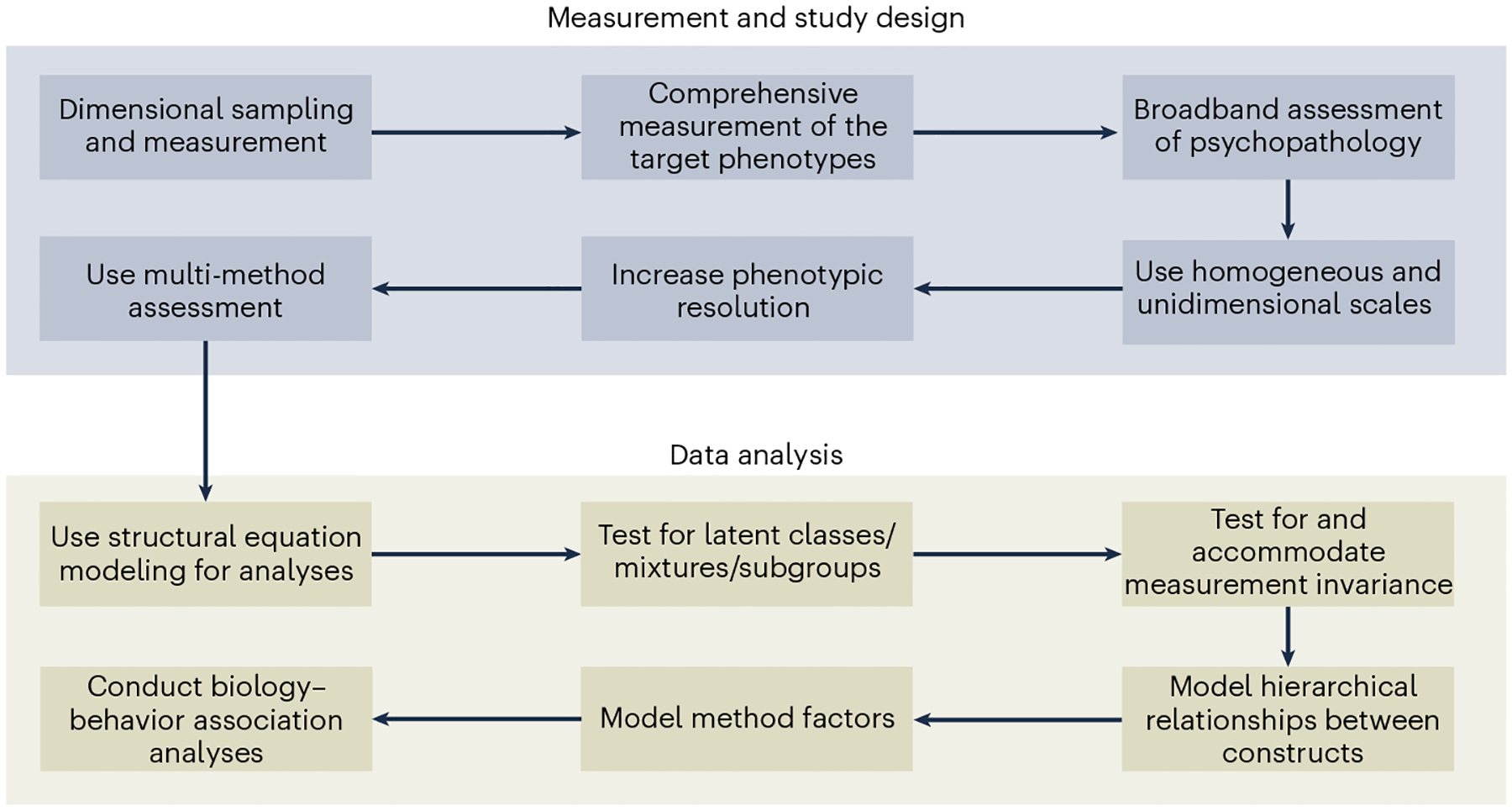
Example workflow for a precision psychiatric phenotyping approach in the context of a biology–psychopathology association study.
Deep phenotyping and use of standardized measures
Existing large-scale databases—such as the UK Biobank65—have a large number of participants who completed an array of measures. However, a limitation of these databases is minimal phenotyping of specific psychopathology phenotypes32. To address problems of minimal and inconsistent phenotyping, we recommend comprehensive assessment using a deep phenotyping approach (comprehensive assessment of one or more phenotypes) with standardized psychopathology measures that can be widely adopted (for example, Box 3), and which are better suited for data pooling via established psychiatric research consortia (for example, ENIGMA and PGC)32. Broadband assessment of multiple dimensions of psychopathology should be undertaken due to the highly comorbid nature of mental health problems64. An advantage of deep phenotyping is that it enables the identification and accommodation of comorbidity, as well as person-centered and variable-centered heterogeneity. Deep phenotyping also facilitates greater comparability across studies and the potential harmonization of datasets. Examples of deep phenotyping can be found in existing cohorts30,31.
Use of homogeneous unidimensional scales and hierarchical modeling
Construct homogeneity (that is, the assumption or evidence that a construct reflects variance in a single phenotype) and unidimensionality (that is, the covariance amongst a homogenous item set is captured by one factor or latent variable, as opposed to two or more factors in the case of multidimensionality) are important qualities of scales used to assess psychopathology that enable researchers to isolate the specific sources of variance associated with biological measures66. Relatedly, owing to the potential empirical overlap of symptom components or empirical syndromes at low levels of the psychopathology hierarchy, it is important that the measures chosen assess homogeneous components with high discriminant validity to avoid redundancy43. We thus advocate for a ‘splitting’ approach in which psychopathological constructs are dissected into finer-grained, lower-order homogeneous constructs to isolate specific variance while taking account of the hierarchical organization of these phenotypes67. A previous study68 provides an example of a splitting approach that identified significant associations between polygenic risk for schizophrenia and psychometric measures of schizotypy in a non-clinical sample that were otherwise obscured by the use of raw scores or a ‘lumping approach’. Unidimensionality of a construct can be evaluated using factor analysis within a structural equation modeling framework (Box 4).
Psychiatric symptoms are intrinsically hierarchical. Even homogeneous scales typically contain sources of variance spanning multiple levels of the hierarchy43. Failure to account for this structure leads to measurement contamination, and reduced reliability and validity for investigating biological associations (compare with example 1 of the Supplementary Information). Phenotypic complexity, multidimensionality, the heterogeneity problem, and the comorbidity problem can all be addressed via hierarchical modeling. There are two approaches to modeling the hierarchical structure of psychopathology: bottom up and top down. Bottom-up approaches leverage higher-order factor models and confirmatory factor analysis within an SEM framework (Box 4), with narrower psychiatric syndromes modeled at the first stage and broader spectra modeled at the second (for a tutorial, see ref. 69). Using a bifactor model, hierarchical sources of variance can be partitioned into a common factor (for example, p-factor) and orthogonal specific factors (for example, internalizing, externalizing; see example 1 of the Supplementary Information for a detailed illustration)44. An alternative bottom-up approach uses hierarchical clustering, where questionnaire items or subscales are organized into homogeneous clusters based on shared features70.
The top-down approach is the bass-ackwards method71. The bass-ackwards method is useful for explicating complex hierarchical structures top down and involves extracting an increasing number of orthogonal principal components to represent the major dimensions of a multi-level hierarchy. The first unrotated principal component captures covariance amongst items or subscales from psychopathology questionnaires at the broadest level. In the second iteration of the method, two orthogonally rotated principal components are extracted; followed by three at the next iteration and so on. Component correlations are calculated between adjacent levels to evaluate continuity versus differentiation of psychopathology components. Proceeding further down the hierarchy, the covariance structure becomes differentiated into dimensions that are increasingly narrow conceptually and empirically, until distinct behavioral syndromes or symptom constellations are isolated. An example of the bass-ackwards method in the ABCD data is provided in ref. 72.
Increasing phenotypic resolution
To address the issue of low phenotypic resolution, items can be carefully selected within an iIRT framework (Box 5) so that they assay psychopathological severity across the full length of the latent-trait continuum, offering psychometric precision at all levels of the measured construct40. Alternatively, it is possible to select measures that have already been optimized within an IRT framework to increase measurement precision across the entire latent-trait continuum (for example, the computerized adaptive assessment of personality disorder; CAT-PD73). For unipolar traits, it is possible to bolster measurement precision with items from a related construct that represents the opposite (that is, adaptive) end of the continuum74. We demonstrate the utility of this approach in example 4 of the Supplementary Information, where we bolster the lower end of the CBCL attention problems latent trait continuum by pooling the items from this scale with items taken from the Early Adolescent Temperament Questionnaire – Revised (EATQ-R)17 effortful control subscale, which measures the adaptive end of the attentional control/attentional problems continuum.
Box 5. Item response theory.
IRT is a sophisticated approach to psychometric scale construction, evaluation and refinement and has been increasingly recommended for, and applied, in psychopathology research100. IRT encapsulates a set of measurement models and statistical methods that can be used to empirically model item level data100. The two-parameter logistic (2PL) model for dichotomous item response data and its extension for polytomous item response data, the graded response (GR) model, are the most commonly used models45,101. Two main parameters of interest are generated through IRT analysis: (1) a slope (also ‘discrimination’) parameter (α); and (2) a threshold (also severity or location) parameter (β). Slope parameters are akin to factor loadings and indicate how well an item measures the latent trait. They are measured in a logistic metric, generally ranging between ±2.8, with higher values indicating that an item is more discriminating between different levels of a latent trait100. Threshold parameters indicate the location on the latent trait continuum where an item is most sensitive to different levels of the latent trait. They are measured in a standardized metric (that is, M = 0, s.d. = 1) generally ranging between ±3, with more extreme values indicating that an item is sensitive to lower and higher levels of symptom severity100. These item-level parameters enable the amount of measurement precision, or ‘information’, to be quantified. Item information is additive and can be combined to represent the total measurement precision of items across the latent-trait continuum47. Information (I) can then be transformed into a standard metric of internal consistency reliability (ref. 101). Items can thus be carefully selected to optimize measurement precision across the whole latent-trait continuum. Furthermore, items with high local dependence (that is, correlated residual variances) can be identified as redundant and removed. Despite the appeal of IRT for optimizing phenotypic precision in psychopathology research, it has not been utilized widely for identifying associations between psychometric constructs and biological measures.
Address measurement non-invariance
Measurement invariance should be thoroughly evaluated across groups, including sex/gender, race/ethnicity and developmental stage. There are multiple resources for invariance testing, including analytic flow charts and checklists50. Differential item function (DIF) testing within an IRT framework provides a powerful approach to invariance testing, but requires larger sample sizes and involves more restrictive assumptions75. Where full invariance does not hold, partial invariance can be considered by freely estimating one or more model parameters in the comparison group76. Alternatively, researchers can utilize Bayesian approximate invariance testing, which is useful when there are many small, trivial differences between group parameters of no substantive interest, but which in combination result in poor model fit76. Groups or subsamples with partial non-invariance of their model parameters can still be meaningfully compared in some circumstances76.
Measurement non-invariance can be accommodated in several ways. Groups or subsamples with fully non-invariant measurement parameters for psychiatric phenotypes should be analyzed separately. It is also possible to circumvent issues of measurement non-equivalence within both factor analytic and IRT frameworks by removing items identified as having non-invariant factor loadings or intercepts, or slope and threshold parameters, to ensure the equivalence of the latent variable across groups. However, in these instances researchers should be cautious of changing the substantive interpretation of the construct by narrowing its scope and breadth (that is, the attenuation paradox).
Mixture modeling
In contrast to situations where subgroups are easily identified and differentiated based on manifest, discrete characteristics such as sex and ethnicity, there are situations where subgroups embedded within the data are not directly observed, resulting in person-centered heterogeneity. Thus, prior to conducting biology–behavior association studies, it is important to verify that the psychiatric phenotypes can be treated as continuous dimensions in the sample. Mixture modeling provides a useful approach for investigating person-centered heterogeneity77. Mixture modeling is a particularly promising approach because it can identify latent classes or clinical subtypes, which often characterize psychopathology phenotypes77. Entropy provides a summary measure of the classification accuracy of participants based on the posterior probabilities of class membership within a mixture modeling analysis. It can range between 0 and 1.00, with higher entropy indicating better classification accuracy. When entropy is high (for example, ≥0.80) class membership can be used as a discrete categorical variable for subsequent analyses to compare results between classes. However, where entropy is low, classes must be compared using alternative analytic approaches that take into account the probabilistic nature of class membership. By identifying and analyzing subtypes, the confounding impact of sample heterogeneity on studies of the associations between biology and psychopathology can be reduced34. In example 5 of the Supplementary Information, we apply mixture modeling to the attention problems CBCL scale, using data from the ABCD 2-year follow-up. Results reveal evidence for two latent classes with different empirical distributions and item response profiles on the CBCL. These observations suggest that failure to account for the latent categorical structure of the attention problems scale could lead to erroneous results in biology–psychopathology association studies.
Multimethod assessment
A fundamental tenet of psychometrics is that measurement of a psychological attribute represents a trait–method unit, combining a person’s true score with systematic measurement error related to the assessment method66. Thus, at least two different assessment methods are required to differentiate the true score for a trait measure from method effects78. The recommended approach to circumventing issues of method bias is to use multimethod assessment and then implement statistical remedies to identify and exclude the method factors and decompose an observed score into true score, method variance (systematic error) and random measurement error60,78. The optimal statistical method for removing method variance is the trait method minus one [T(M-1)] model estimated within an SEM framework (Box 4)79.
In example 6 of the Supplementary Information, we apply the T(M-1) method to the new composite scale we constructed in example 4, which combined CBCL attention problems scale items and the EATQ-R effortful control subscale items of the ABCD data. The purpose of applying the T(M-1) model was to control for method variance associated with subjective report by the primary caregivers and in doing so increase signal-to-noise ratio. To do so, we incorporated neurocognitive measures of the target attention problems construct; specifically, stop signal reaction time from the stop signal task and d-prime as an estimate of working memory from the n-back task, both of which are well-established endophenotypes of ADHD80,81. We were then able to specify the neurocognitive measures as the reference method, such that loadings from the CBCL and EATQ-R caregiver report items on the target attention problems factor captured only that variance shared with the neurocognitive measures. A methods factor captured the residual variance in subjective report by the primary caregivers that was unique to these measures79. We found that the attention problems factor was associated with polygenic risk for ADHD. By contrast, the methods factor that captured variance specific to caregiver-report measures of attention problems and attention control abilities was not significantly related to polygenic risk for ADHD (Supplementary Fig. 27). Thus, the T(M-1) model yielded a genetic association that was otherwise obscured by standard analyses.
Conclusions
It has been suggested that large, consortia-sized samples are necessary to discover robust and reproducible biology–psychopathology associations. Larger sample sizes are not sufficient to resolve the issues introduced by imprecise or otherwise suboptimal psychiatric phenotypes. As a field, we must first improve our measurement techniques. We recommended broadband, transdiagnostic assessment of hierarchically organized, unidimensional and homogeneous psychopathology dimensions across the full range of the severity spectrum. We encourage greater focus on deep phenotyping, measurement invariance, phenotypic resolution, and person-centered and variable-centered heterogeneity. A voluminous psychometrics literature—and the worked examples featured in this Review—make clear that this multi-faceted strategy will increase validity, reliability, effect sizes, statistical power and, ultimately, replicability.
Supplementary Material
Acknowledgements
J.T. was supported by a Turner impact fellowship from the Turner Institute for Brain and Mental Health, Monash University, Australia. A.F. was supported by the Sylvia and Charles Viertel Foundation, the National Health and Medical Research Council (grant numbers: 1146292 and 1197431) and the Australian Research Council (grant number: DP200103509). A.J.S. was supported by the National Institute of Mental Health (grant numbers: AA030042, MH131264 and MH121409) and the University of Maryland.
Footnotes
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s44220-023-00057-5.
References
- 1.Perkins ER, Latzman RD & Patrick CJ Interfacing neural constructs with the Hierarchical Taxonomy of Psychopathology: ‘why’ and ‘how’. Personal. Ment. Health 14, 106–122 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Insel T et al. Research Domain Criteria (RDoC): toward a new classification framework for research on mental disorders. Am. J. Psychiatry 167, 748–751 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Hyman SE Can neuroscience be integrated into the DSM-V? Nat. Rev. Neurosci 8, 725–732 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Singh I & Rose N Biomarkers in psychiatry. Nature 460, 202–207 (2009). [DOI] [PubMed] [Google Scholar]
- 5.First MB et al. Clinical applications of neuroimaging in psychiatric disorders. Am. J. Psychiatry 175, 915–916 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marek S et al. Reproducible brain-wide association studies require thousands of individuals. Nature 603, 654–660 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poldrack RA et al. Scanning the horizon: towards transparent and reproducible neuroimaging research. Nat. Rev. Neurosci 18, 115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saggar M & Uddin LQ Pushing the boundaries of psychiatric neuroimaging to ground diagnosis in biology. eNeuro 10.1523/eneuro.0384-19.2019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sha Z, Wager TD, Mechelli A & He Y Common dysfunction of large-scale neurocognitive networks across psychiatric disorders. Biol. Psychiatry 85, 379–388 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Smoller JW et al. Psychiatric genetics and the structure of psychopathology. Mol. Psychiatry 24, 409–420 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nour MM, Liu Y & Dolan RJ Functional neuroimaging in psychiatry and the case for failing better. Neuron 110, 2524–2544 (2022). [DOI] [PubMed] [Google Scholar]
- 12.Sullivan PF et al. Psychiatric genomics: an update and an agenda. Am. J. Psychiatry 175, 15–27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kundu P, Inati SJ, Evans JW, Luh W-M & Bandettini PA Differentiating BOLD and non-BOLD signals in fMRI time series using multi-echo EPI. NeuroImage 60, 1759–1770 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parkes L, Fulcher B, Yücel M & Fornito A An evaluation of the efficacy, reliability, and sensitivity of motion correction strategies for resting-state functional MRI. NeuroImage 171, 415–436 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Kong R et al. Individual-specific areal-level parcellations improve functional connectivity prediction of behavior. Cereb. Cortex 31, 4477–4500 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Visscher PM et al. 10 years of GWAS discovery: biology, function, and ranslation. Am. J. Hum. Genet 101, 5–22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Volkow ND et al. The conception of the ABCD study: from substance use to a broad NIH collaboration. Dev. Cogn. Neurosci 32, 4–7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lilienfeld SO The Research Domain Criteria (RDoC): an analysis of methodological and conceptual challenges. Behav. Res. Ther 62, 129–139 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Xing X-X & Zuo X-N The anatomy of reliability: a must read for future human brain mapping. Sci. Bull 63, 1606–1607 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Zuo XN, Xu T & Milham MP Harnessing reliability for neuroscience research. Nat. Hum. Behav 3, 768–771 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Nikolaidis A et al. Suboptimal phenotypic reliability impedes reproducible human neuroscience. Preprint at bioRxiv 10.1101/2022.07.22.501193 (2022). [DOI] [Google Scholar]
- 22.Falk EB et al. What is a representative brain? Neuroscience meets population science. Proc. Natl. Acad. Sci. USA 110, 17615–17622 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Markon KE, Chmielewski M & Miller CJ The reliability and validity of discrete and continuous measures of psychopathology: a quantitative review. Psychol. Bull 137, 856–879 (2011). [DOI] [PubMed] [Google Scholar]
- 24.van der Sluis S, Posthuma D, Nivard MG, Verhage M & Dolan CV Power in GWAS: lifting the curse of the clinical cut-off. Mol. Psychiatry 18, 2–3 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Fisher JE, Guha A, Heller W & Miller GA Extreme-groups designs in studies of dimensional phenomena: Advantages, caveats, and recommendations. J. Abnorm. Psychol 129, 14–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Angold A, Costello EJ, Farmer EMZ, Burns BJ & Erkanli A Impaired but undiagnosed. J. Am. Acad. Child Adolesc. Psychiatry 38, 129–137 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Preacher KJ in Extreme Groups Designs in the Encyclopedia of Clinical Psychology Vol. 2 (eds. Cautin RL & Lilienfeld SO) 1189–1192 (John Wiley and Sons, 2015). [Google Scholar]
- 28.Dong H-M et al. Charting brain growth in tandem with brain templates at school age. Sci. Bull 65, 1924–1934 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Peterson RE et al. Genome-wide association studies in ancestrally diverse populations: opportunities, methods, pitfalls, and recommendations. Cell 179, 589–603 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu S et al. Chinese color nest project: an accelerated longitudinal brain-mind cohort. Dev. Cogn. Neurosci 52, 101020 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tobe RH et al. A longitudinal resource for studying connectome development and its psychiatric associations during childhood. Sci. Data 9, 300 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanchez-Roige S & Palmer AA Emerging phenotyping strategies will advance our understanding of psychiatric genetics. Nat. Neurosci 23, 475–480 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newson JJ, Hunter D & Thiagarajan TC The heterogeneity of mental health assessment. Front. Psychiatry 10.3389/fpsyt.2020.00076 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feczko E et al. The heterogeneity problem: approaches to identify psychiatric subtypes. Trends Cogn. Sci 23, 584–601 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schnack HG & Kahn RS Detecting neuroimaging biomarkers for psychiatric disorders: sample size matters. Front. Psychiatry 7, 50 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Z et al. Brain network informed subject community detection in early-onset schizophrenia. Sci. Rep 4, 5549 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hodgson K, McGuffin P & Lewis CM Advancing psychiatric genetics through dissecting heterogeneity. Hum. Mol. Genet 26, R160–R165 (2017). [DOI] [PubMed] [Google Scholar]
- 38.De Nadai AS, Hu Y & Thompson WK Data pollution in neuropsychiatry—an under-recognized but critical barrier to research progress. JAMA Psychiatry 79, 97–98 (2022). [DOI] [PubMed] [Google Scholar]
- 39.Reise SP, Bonifay WE & Haviland MG Scoring and modeling psychological measures in the presence of multidimensionality. J. Pers. Assess 95, 129–140 (2013). [DOI] [PubMed] [Google Scholar]
- 40.van der Sluis S, Verhage M, Posthuma D & Dolan CV Phenotypic complexity, measurement bias, and poor phenotypic resolution contribute to the missing heritability problem in genetic association studies. PLoS ONE 5, e13929 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caspi A & Moffitt TE All for one and one for all: mental disorders in one dimension. Am. J. Psychiatry 175, 831–844 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kotov R et al. The Hierarchical Taxonomy of Psychopathology (HiTOP): a quantitative nosology based on consensus of evidence. Annu. Rev. Clin. Psychol 17, 83–108 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Clark LA & Watson D Constructing validity: new developments in creating objective measuring instruments. Psychol. Assess 31, 1412–1427 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reise SP The rediscovery of bifactor measurement models. Multivariate Behav. Res 47, 667–696 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reise SP & Waller NG Item response theory and clinical measurement. Annu. Rev. Clin. Psychol 5, 27–48 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Rosopa PJ, Schaffer MM & Schroeder AN Managing heteroscedasticity in general linear models. Psychol. Methods 18, 335–351 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Thomas ML The value of item response theory in clinical assessment: a review. Assessment 18, 291–307 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Streiner DL Starting at the beginning: an introduction to coefficient alpha and internal consistency. J. Pers. Assess 80, 99–103 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Saccenti E, Hendriks MHWB & Smilde AK Corruption of the Pearson correlation coefficient by measurement error and its estimation, bias, and correction under different error models. Sci. Rep 10, 438 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vandenberg RJ & Lance CE A review and synthesis of the measurement invariance literature: suggestions, practices, and recommendations for organizational research. Organ. Res. Methods 3, 4–70 (2000). [Google Scholar]
- 51.Miettunen J, Nordstrom T, Kaakinen M & Ahmed AO Latent variable mixture modeling in psychiatric research: a review and application. Psychol. Med 46, 457–467 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Achenbach TM The Achenbach System of Empirically Based Assessment (ASEBA): Development, Findings, Theory, and Applications (University of Vermont, Research Center for Children, Youth and Families, 2009). [Google Scholar]
- 53.Kelly EL Interpretation of Educational Measurements (World Book, 1927). [Google Scholar]
- 54.Fried EI Moving forward: how depression heterogeneity hinders progress in treatment and research. Expert Rev. Neurother 17, 423–425 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Fried EI & Nesse RM Depression is not a consistent syndrome: an investigation of unique symptom patterns in the STAR*D study. J. Affect. Disord 172, 96–102 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wager TD & Woo C-W Imaging biomarkers and biotypes for depression. Nat. Med 23, 16–17 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Fried EI & Nesse RM Depression sum-scores don’t add up: why analyzing specific depression symptoms is essential. BMC Med. 13, 72 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kendler KS, Aggen SH & Neale MC Evidence for multiple genetic factors underlying DSM-IV criteria for major depression. JAMA Psychiatry 70, 599–607 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Podsakoff PM, MacKenzie SB, Lee J & Podsakoff NP Common method biases in behavioral research: a critical review of the literature and recommended remedies. J. Appl. Psychol 88, 879–903 (2003). [DOI] [PubMed] [Google Scholar]
- 60.Podsakoff PM, MacKenzie SB & Podsakoff NP Sources of method bias in social science research and recommendations on how to control it. Annu. Rev. Psychol 63, 539–569 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Haslam N, Holland E & Kuppens P Categories versus dimensions in personality and psychopathology: a quantitative review of taxometric research. Psychol. Med 42, 903–920 (2012). [DOI] [PubMed] [Google Scholar]
- 62.Plomin R, Haworth CM & Davis OS Common disorders are quantitative traits. Nat. Rev. Genet 10, 872–878 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Cuthbert BN The RDoC framework: facilitating transition from ICD/DSM to dimensional approaches that integrate neuroscience and psychopathology. World Psychiatry 13, 28–35 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stanton K, McDonnell CG, Hayden EP & Watson D Transdiagnostic approaches to psychopathology measurement: Recommendations for measure selection, data analysis, and participant recruitment. J. Abnorm. Psychol 129, 21–28 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Allen N et al. UK Biobank: current status and what it means for epidemiology. Health Policy Technol. 1, 123–126 (2012). [Google Scholar]
- 66.Strauss ME & Smith GT Construct validity: advances in theory and methodology. Annu. Rev. Clin. Psychol 5, 1–25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Karcher NR, Michelini G, Kotov R & Barch DM Associations between resting-state functional connectivity and a hierarchical dimensional structure of psychopathology in middle childhood. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 6, 508–517 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tiego J et al. Dissecting schizotypy and its association with cognition and polygenic risk for schizophrenia in a nonclinical sample. Schizophr Bull. 10.1093/schbul/sbac016 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Conway CC, Forbes MK & South SC A Hierarchical Taxonomy of Psychopathology (HiTOP) primer for mental health researchers. Clin. Psychol. Sci 10.1177/21677026211017834 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yim O & Ramdeen KT Hierarchical cluster analysis: comparison of three linkage measures and application to psychological data. Quant. Methods Psychol 11, 8–21 (2015). [Google Scholar]
- 71.Goldberg LR Doing it all bass-ackwards: the development of hierarchical factor structures from the top down. J. Res. Pers 40, 347–358 (2006). [Google Scholar]
- 72.Michelini G et al. Delineating and validating higher-order dimensions of psychopathology in the Adolescent Brain Cognitive Development (ABCD) study. Transl. Psychiatry 9, 261 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Simms LJ et al. Computerized adaptive assessment of personality disorder: introducing the CAT–PD project. J. Pers. Assess 93, 380–389 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Greven CU, Buitelaar JK & Salum GA From positive psychology to psychopathology: the continuum of attention‐deficit hyperactivity disorder. J. Child Psychol. Psychiatry 59, 203–212 (2018). [DOI] [PubMed] [Google Scholar]
- 75.Stark S, Chernyshenko OS & Drasgow F Detecting differential item functioning with confirmatory factor analysis and item response theory: toward a unified strategy. J. Appl. Psychol 91, 1292–1306 (2006). [DOI] [PubMed] [Google Scholar]
- 76.van de Schoot R et al. Facing off with Scylla and Charybdis: a comparison of scalar, partial, and the novel possibility of approximate measurement invariance. Front. Psychol 4, 770 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Clark SL et al. Models and strategies for factor mixture analysis: an example concerning the structure underlying psychological disorders. Struct. Equation Modell 20, 681–703 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Eid M, Lischetzke T, Nussbeck FW & Trierweiler LI Separating trait effects from trait-specific method effects in multitrait-multimethod models: a multiple-indicator CT-C(M-1) model. Psychol. Methods 8, 38–60 (2003). [DOI] [PubMed] [Google Scholar]
- 79.Eid M, Geiser C & Koch T Measuring method effects: from traditional to design-oriented approaches. Curr. Dir. Psychol. Sci 25, 275–280 (2016). [Google Scholar]
- 80.Aron AR & Poldrack RA The cognitive neuroscience of response inhibition: relevance for genetic research in attention-deficit/hyperactivity disorder. Biol. Psychiatry 57, 1285–1292 (2005). [DOI] [PubMed] [Google Scholar]
- 81.Martinussen R, Hayden J, Hogg-Johnson S & Tannock R A meta-analysis of working memory impairments in children with attention-deficit/hyperactivity disorder. J. Am. Acad. Child Adolesc. Psychiatry 44, 377–384 (2005). [DOI] [PubMed] [Google Scholar]
- 82.DeVellis RF Classical test theory. Med. Care 44, S50–S59 (2006). [DOI] [PubMed] [Google Scholar]
- 83.Nunnally JC & Bernstein I Psychometric Theory 3rd edn (McGraw-Hill, 1994). [Google Scholar]
- 84.Antonakis J, Bendahan S, Jacquart P & Lalive R On making causal claims: a review and recommendations. Leadersh. Q 21, 1086–1120 (2010). [Google Scholar]
- 85.Kendell R & Jablensky R Distinguishing between the validity and utility of psychiatric diagnoses. Am. J. Psychiatry 160, 4–12 (2003). [DOI] [PubMed] [Google Scholar]
- 86.Kendler KS The phenomenology of major depression and the representativeness and nature of DSM criteria. Am. J. Psychiatry 173, 771–780 (2016). [DOI] [PubMed] [Google Scholar]
- 87.Kotov R, Ruggero CJ, Krueger RF, Watson D & Zimmerman M The perils of hierarchical exclusion rules: a further word of caution. Depress. Anxiety 35, 903–904 (2018). [DOI] [PubMed] [Google Scholar]
- 88.Caspi A et al. The p factor: one general psychopathology factor in the structure of psychiatric disorders? Clin. Psychol. Sci 2, 119–137 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Allsopp K, Read J, Corcoran R & Kinderman P Heterogeneity in psychiatric diagnostic classification. Psychiatry Res. 279, 15–22 (2019). [DOI] [PubMed] [Google Scholar]
- 90.Cuthbert BN & Insel TR Toward the future of psychiatric diagnosis: the seven pillars of RDoC. BMC Med. 11, 126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Simms LJ et al. Development of measures for the hierarchical taxonomy of psychopathology (HiTOP): a collaborative scale development project. Assessment 29, 3–16 (2021). [DOI] [PubMed] [Google Scholar]
- 92.HiTOP Friendly Measures. HiTOP Clinical Network https://hitop.unt.edu/clinical-tools/hitop-friendly-measures (accessed 1 October 2022).
- 93.Lahey BB, Krueger RF, Rathouz PJ, Waldman ID & Zald DH A hierarchical causal taxonomy of psychopathology across the life span. Psychol. Bull 143, 142–186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Michelini G, Palumbo IM, DeYoung CG, Latzman RD & Kotov R Linking RDoC and HiTOP: a new interface for advancing psychiatric nosology and neuroscience. Clin. Psychol. Rev 86, 102025 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hair JF, Black WC, Babin BJ & Anderson RE Multivariate Data Analysis 7th edn (Pearson Education, 2014). [Google Scholar]
- 96.Grice JW Computing and evaluating factor scores. Psychol. Methods 6, 430–450 (2001). [PubMed] [Google Scholar]
- 97.Devlieger I, Mayer A & Rosseel Y Hypothesis testing using factor score regression: a comparison of four methods. Educ. Psychol. Meas 76, 741–770 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim J, Zhu W, Chang L, Bentler PM & Ernst T Unified structural equation modeling approach for the analysis of multisubject, multivariate functional MRI data. Hum. Brain Mapp 28, 85–93 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kline RB Principles and Practice of Structural Equation Modeling 4th edn (Guilford, 2015). [Google Scholar]
- 100.Reise SP & Rodriguez A Item response theory and the measurement of psychiatric constructs: some empirical and conceptual issues and challenges. Psychol. Med 46, 2025–2039 (2016). [DOI] [PubMed] [Google Scholar]
- 101.de Ayala RJ The Theory and Practice of Item Response Theory (Guilford, 2009). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


